28.3 Amino Acids, Proteins, and Enzymes
Learning Objectives
By the end of this section, you will be able to:
- Recognize amino acids and classify them based on the characteristics of their side chains
- Explain how an amino acid can act as both an acid and a base
- Explain how a peptide is formed from individual amino acids
- Explain why the sequence of amino acids in a protein is important
- Describe the four levels of protein structure.
- Identify the types of attractive interactions that hold proteins in their most stable three-dimensional structure.
- Explain what happens when proteins are denatured.
- Explain the functions of enzymes and how enzymes are classified and named
- Describe the interaction between an enzyme and its substrate
From Structural Integrity to Biological Function
Proteins may be defined as compounds of high molar mass consisting largely or entirely of chains of amino acids. Their masses range from several thousand to several million daltons (Da). In addition to carbon, hydrogen, and oxygen atoms, all proteins contain nitrogen and sulfur atoms, and many also contain phosphorus atoms and traces of other elements. Proteins serve a variety of roles in living organisms and are often classified by these biological roles. Muscle tissue is largely protein, as are skin and hair. Proteins are present in the blood, in the brain, and even in tooth enamel. Each type of cell in our bodies makes its own specialized proteins, as well as proteins common to all or most cells. We begin our study of proteins by looking at the properties and reactions of amino acids, which is followed by a discussion of how amino acids link covalently to form peptides and proteins. We end the chapter with a discussion of enzymes—the proteins that act as catalysts in the body.
Spotlight on Everyday Chemistry: 1923 Nobel Prize
The 1923 Nobel Prize in Medicine or Physiology was awarded to Frederick Grant Banting and John James Richard Macleod for their discovery of the protein insulin. In 1958, the Nobel Prize in Chemistry was awarded to Frederick Sanger for his discoveries concerning the structure of proteins and, in particular, the structure of insulin. What is so important about insulin that two Nobel Prizes have been awarded for work on this protein?
Insulin is a hormone that is synthesized in the pancreas. Insulin stimulates the transport of glucose into cells throughout the body and the storage of glucose as glycogen. People with diabetes do not produce insulin or use it properly. The isolation of insulin in 1921 led to the first effective treatment for these individuals.
Amino Acids
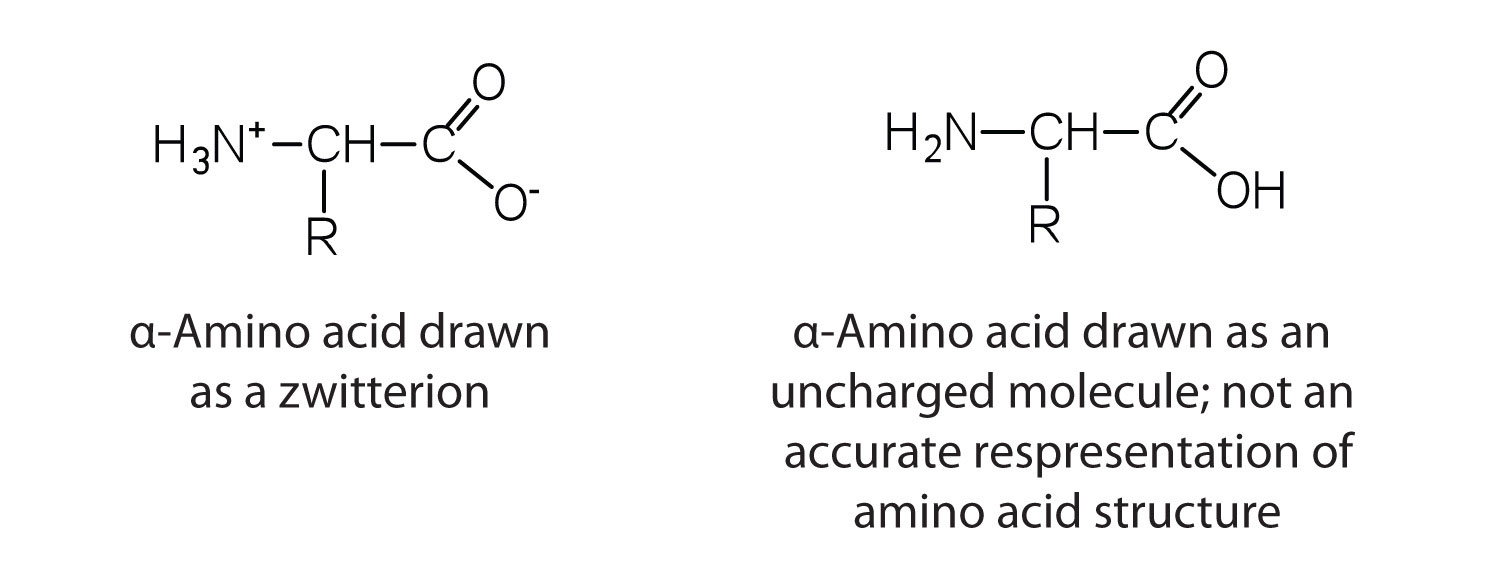
The proteins in all living species, from bacteria to humans, are constructed from the same set of 20 amino acids, so called because each contains an amino group attached to a carboxylic acid. The amino acids in proteins are α-amino acids, which means the amino group is attached to the α-carbon of the carboxylic acid. Humans can synthesize only about half of the needed amino acids; the remainder must be obtained from the diet and are known as essential amino acids. However, two additional amino acids have been found in limited quantities in proteins: Selenocysteine was discovered in 1986, while pyrrolysine was discovered in 2002.
The amino acids are colourless, nonvolatile, crystalline solids, melting and decomposing at temperatures above 200°C. These melting temperatures are more like those of inorganic salts than those of amines or organic acids and indicate that the structures of the amino acids in the solid state and in neutral solution are best represented as having both a negatively charged group and a positively charged group. Such a species is known as a zwitterion (Figure 28.3b.).
Classification
In addition to the amino and carboxyl groups, amino acids have a side chain or R group attached to the α-carbon. Each amino acid has unique characteristics arising from the size, shape, solubility, and ionization properties of its R group. As a result, the side chains of amino acids exert a profound effect on the structure and biological activity of proteins. Although amino acids can be classified in various ways, one common approach is to classify them according to whether the functional group on the side chain at neutral pH is nonpolar, polar but uncharged, negatively charged, or positively charged. The structures and names of the 20 amino acids, their one- and three-letter abbreviations, and some of their distinctive features are given in Table 28.3a-d.
| Common Name | Abbreviation | Structural Formula (at pH 6) | Molar Mass | Distinctive Feature |
|---|---|---|---|---|
| glycine | gly (G) | 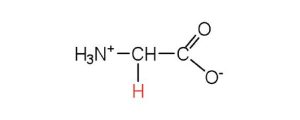 |
75 | the only amino acid lacking a chiral carbon |
| alanine | ala (A) | 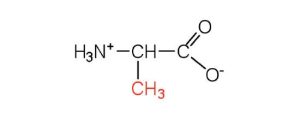 |
89 | — |
| valine | val (V) | 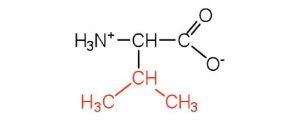 |
117 | a branched-chain amino acid |
| leucine | leu (L) | 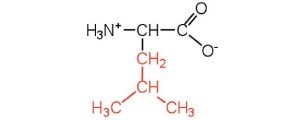 |
131 | a branched-chain amino acid |
| isoleucine | ile (I) | 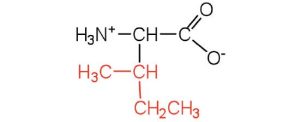 |
131 | an essential amino acid because most animals cannot synthesize branched-chain amino acids |
| phenylalanine | phe (F) | 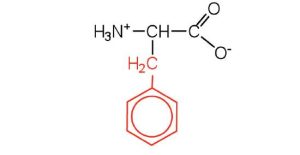 |
165 | also classified as an aromatic amino acid |
| tryptophan | trp (W) | 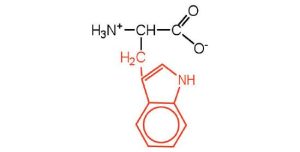 |
204 | also classified as an aromatic amino acid |
| methionine | met (M) | 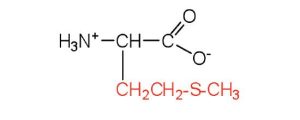 |
149 | side chain functions as a methyl group donor |
| proline | pro (P) | 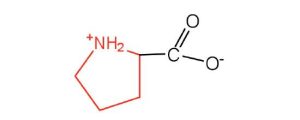 |
115 | contains a secondary amine group; referred to as an α-imino acid |
| Common Name | Abbreviation | Structural Formula (at pH 6) | Molar Mass | Distinctive Feature |
|---|---|---|---|---|
| serine | ser (S) | 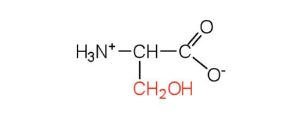 |
105 | found at the active site of many enzymes |
| threonine | thr (T) | 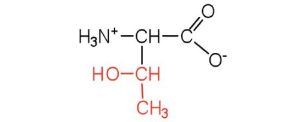 |
119 | named for its similarity to the sugar threose |
| cysteine | cys (C) | 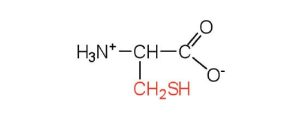 |
121 | oxidation of two cysteine molecules yields cystine |
| tyrosine | tyr (Y) | 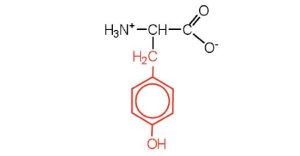 |
181 | also classified as an aromatic amino acid |
| asparagine | asn (N) | 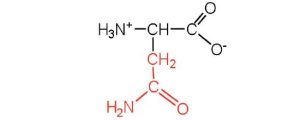 |
132 | the amide of aspartic acid |
| glutamine | gln (Q) | 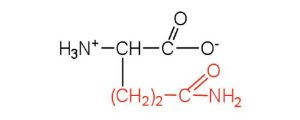 |
146 | the amide of glutamic acid |
| Common Name | Abbreviation | Structural Formula (at pH 6) | Molar Mass | Distinctive Feature |
|---|---|---|---|---|
| aspartic acid | asp (D) | 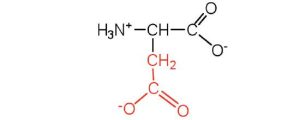 |
132 | carboxyl groups are ionized at physiological pH; also known as aspartate |
| glutamic acid | glu (E) | 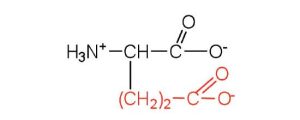 |
146 | carboxyl groups are ionized at physiological pH; also known as glutamate |
| Common Name | Abbreviation | Structural Formula (at pH 6) | Molar Mass | Distinctive Feature |
|---|---|---|---|---|
| histidine | his (H) | 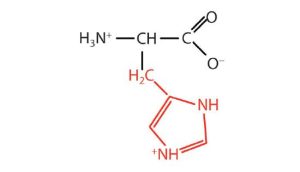 |
155 | the only amino acid whose R group has a pKa (6.0) near physiological pH |
| lysine | lys (K) | 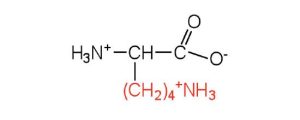 |
147 | — |
| arginine | arg (R) | 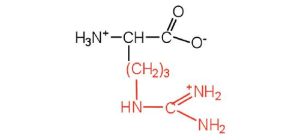 |
175 | almost as strong a base as sodium hydroxide |
Link to Enhanced Learning
Exercise 28.3a
Examine the structures in Table 28.3a.
- For the “Amino acids with a nonpolar R group”, identify the main functional groups in the R section and explain why each is non-polar.
- For the “Amino acids with a polar but neutral R group”, identify the main functional groups in the R section and explain why each is polar.
Check Your Answers:[1]
Source: Exercise 28.3a by Samantha Sullivan Sauer, licensed under CC BY-NC 4.0
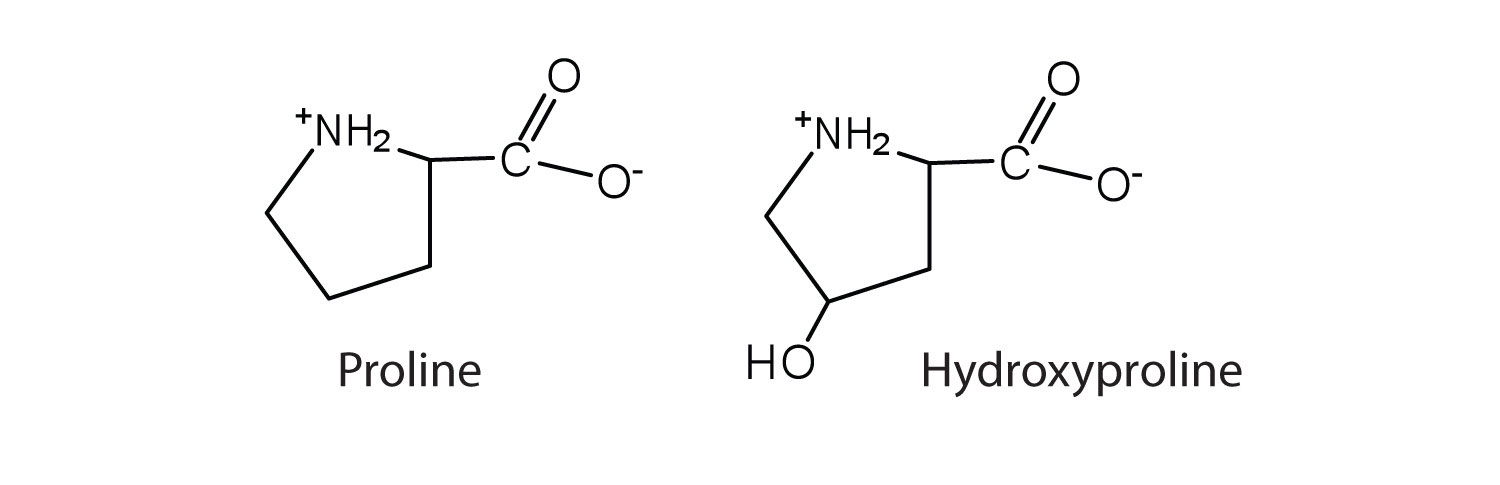
The first amino acid to be isolated was asparagine in 1806. It was obtained from protein found in asparagus juice (hence the name). Glycine, the major amino acid found in gelatin, was named for its sweet taste (Greek glykys, meaning “sweet”). In some cases an amino acid found in a protein is actually a derivative of one of the common 20 amino acids (one such derivative is hydroxyproline, Figure 28.3c.). The modification occurs after the amino acid has been assembled into a protein.
Configuration
Notice in Table 28.3a. that glycine is the only amino acid whose α-carbon is not chiral. Therefore, with the exception of glycine, the amino acids could theoretically exist in either the D- or the L-enantiomeric form and rotate plane-polarized light. As with sugars, chemists used L-glyceraldehyde as the reference compound for the assignment of absolute configuration to amino acids (Figure 28.3d.). Its structure closely resembles an amino acid structure except that in the latter, an amino group takes the place of the OH group on the chiral carbon of the L-glyceraldehyde and a carboxylic acid replaces the aldehyde. Modern stereochemistry assignments using the Cahn-Ingold-Prelog priority rules used ubiquitously in chemistry show that all of the naturally occurring chiral amino acids are S except cysteine which is R.
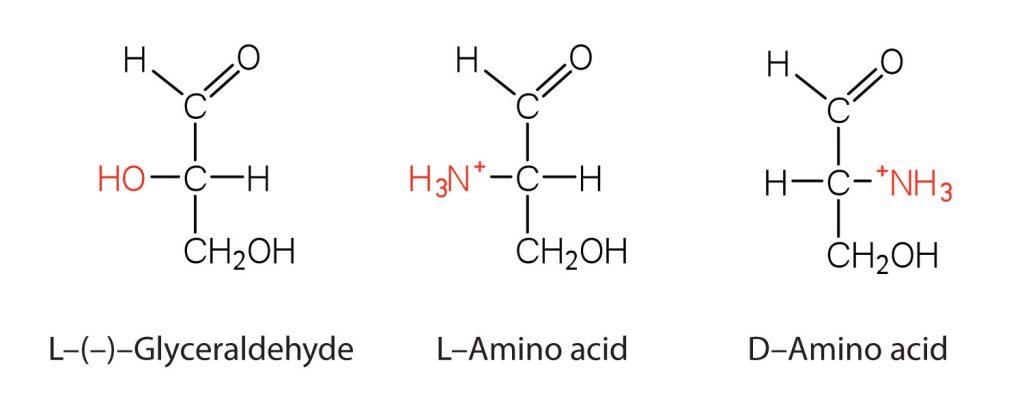
We learned that all naturally occurring sugars belong to the D series. It is interesting, therefore, that nearly all known plant and animal proteins are composed entirely of L-amino acids. However, certain bacteria contain D-amino acids in their cell walls, and several antibiotics (e.g., actinomycin D and the gramicidins) contain varying amounts of D-leucine, D-phenylalanine, and D-valine.
Reactions of Amino Acids
The structure of an amino acid allows it to act as both an acid and a base. An amino acid has this ability because at a certain pH value (different for each amino acid) nearly all the amino acid molecules exist as zwitterions. If acid is added to a solution containing the zwitterion, the carboxylate group captures a hydrogen (H+) ion, and the amino acid becomes positively charged. If base is added, ion removal of the H+ ion from the amino group of the zwitterion produces a negatively charged amino acid. In both circumstances (Figure 28.3e.), the amino acid acts to maintain the pH of the system—that is, to remove the added acid (H+) or base (OH−) from solution.
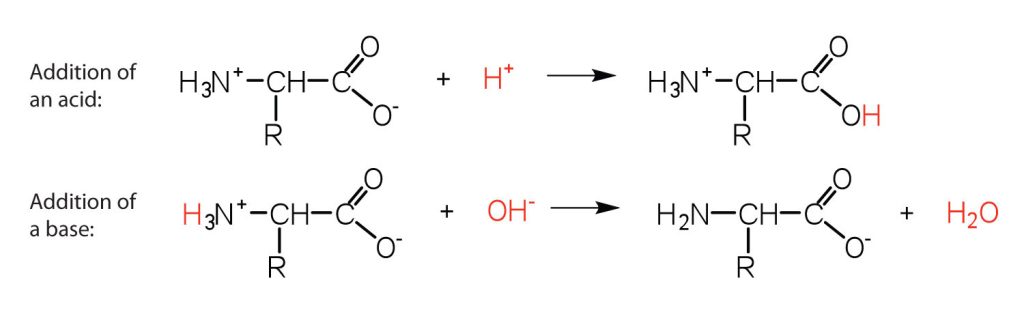
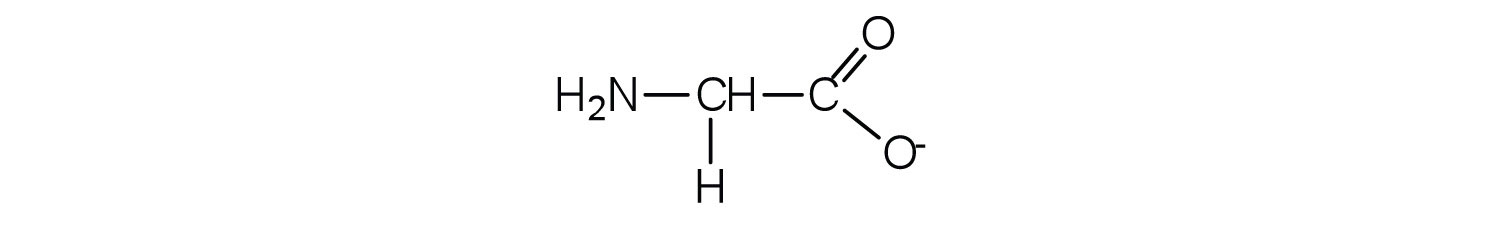
- Draw the structure for the anion formed when glycine (at neutral pH) reacts with a base.
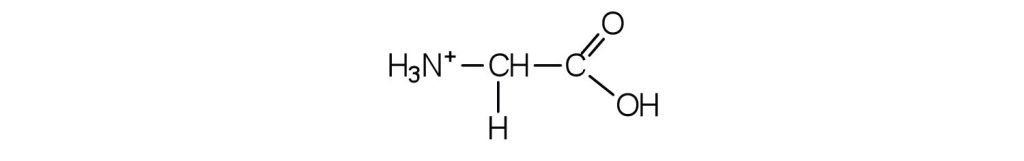
- Draw the structure for the cation formed when glycine (at neutral pH) reacts with an acid.
Solution
- The base removes H+ from the protonated amine group.
b. The acid adds H+ to the carboxylate group.
Example & image source: Introduction to Chemistry: GOB (v. 1.0), CC BY-NC-SA 3.0
The particular pH at which a given amino acid exists in solution as a zwitterion is called the isoelectric point (pI). At its pI, the positive and negative charges on the amino acid balance, and the molecule as a whole is electrically neutral. The amino acids whose side chains are always neutral have isoelectric points ranging from 5.0 to 6.5. The basic amino acids (which have positively charged side chains at neutral pH) have relatively high examples. Acidic amino acids (which have negatively charged side chains at neutral pH) have quite low examples (Table 28.3e.).
| Amino Acid | Classification | pI |
|---|---|---|
| alanine | nonpolar | 6.0 |
| valine | nonpolar | 6.0 |
| serine | polar, uncharged | 5.7 |
| threonine | polar, uncharged | 6.5 |
| arginine | positively charged (basic) | 10.8 |
| histidine | positively charged (basic) | 7.6 |
| lysine | positively charged (basic) | 9.8 |
| aspartic acid | negatively charged (acidic) | 3.0 |
| glutamic acid | negatively charged (acidic) | 3.2 |
Source: “18.2: Reactions of Amino Acids” In Basics of GOB Chemistry (Ball et al.), CC BY-NC-SA 4.0.
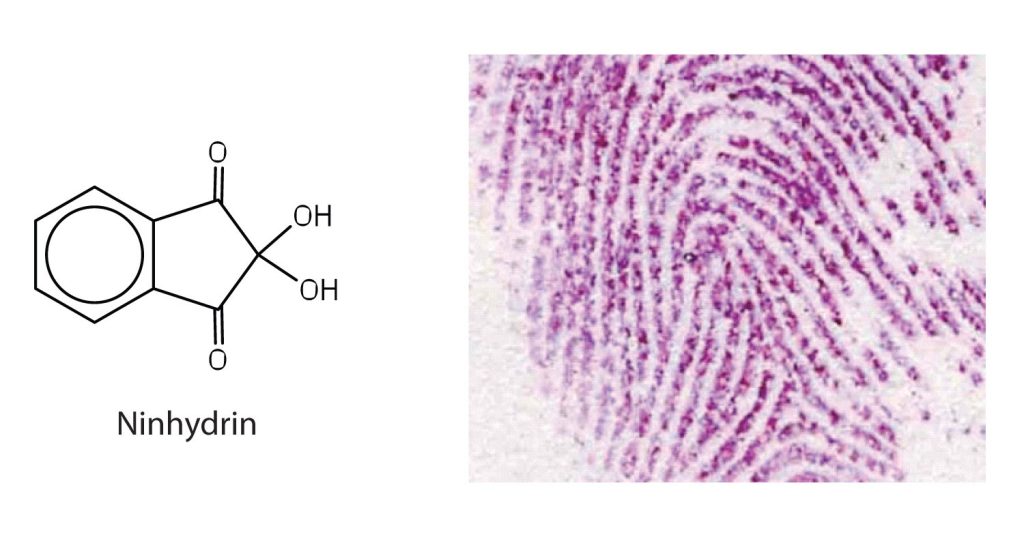
Amino acids undergo reactions characteristic of carboxylic acids and amines. The reactivity of these functional groups is particularly important in linking amino acids together to form peptides and proteins, as you will see later in this chapter. Simple chemical tests that are used to detect amino acids take advantage of the reactivity of these functional groups. An example is the ninhydrin test in which the amine functional group of α-amino acids reacts with ninhydrin to form purple-coloured compounds. Ninhydrin is used to detect fingerprints because it reacts with amino acids from the proteins in skin cells transferred to the surface by the individual leaving the fingerprint (Figure 28.3f.).
Peptides
Two or more amino acids can join together into chains called peptides. We have discussed the reaction between ammonia and a carboxylic acid to form an amide. In a similar reaction, the amino group on one amino acid molecule reacts with the carboxyl group on another, releasing a molecule of water and forming an amide linkage, as seen in Figure 28.3g.
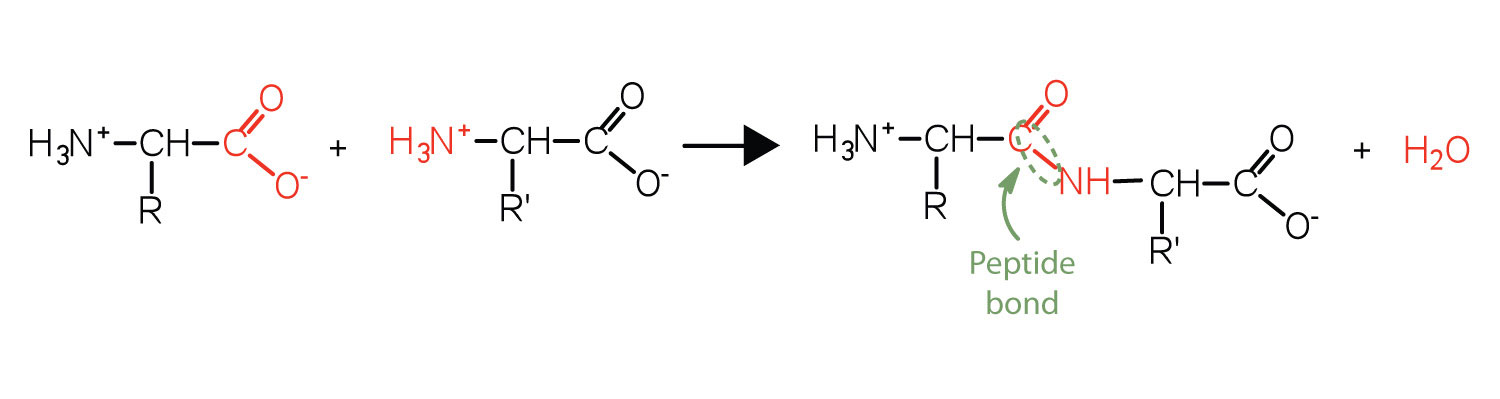
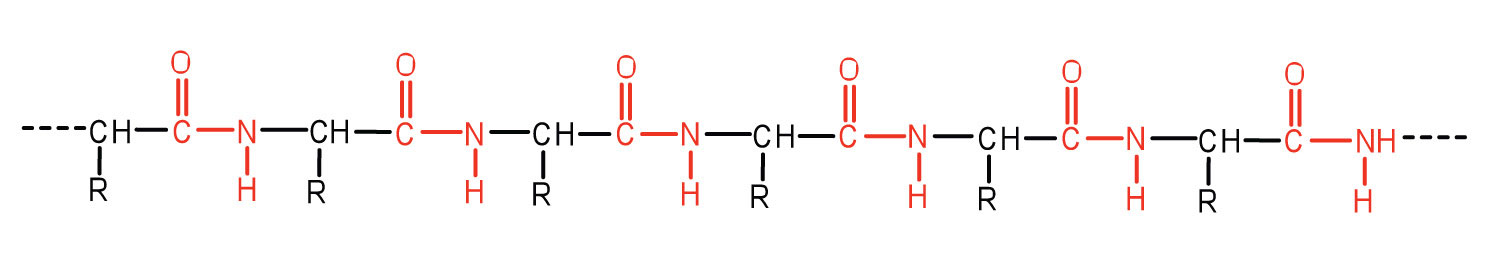
An amide bond joining two amino acid units is called a peptide bond. Note that the product molecule still has a reactive amino group on the left and a reactive carboxyl group on the right. These can react with additional amino acids to lengthen the peptide. The process can continue until thousands of units have joined, resulting in large proteins, as seen in Figure 28.3h.
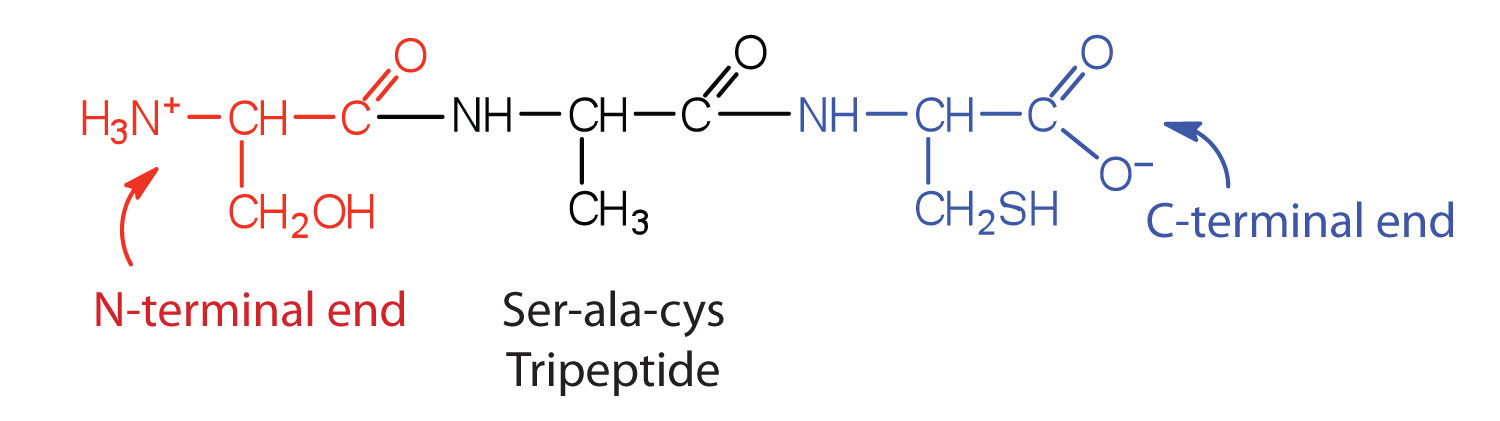
A chain consisting of only two amino acid units is called a dipeptide; a chain consisting of three is a tripeptide. By convention, peptide and protein structures are depicted with the amino acid whose amino group is free (the N-terminal end) on the left and the amino acid with a free carboxyl group (the C-terminal end) to the right, as shown in Figure 28.3i.
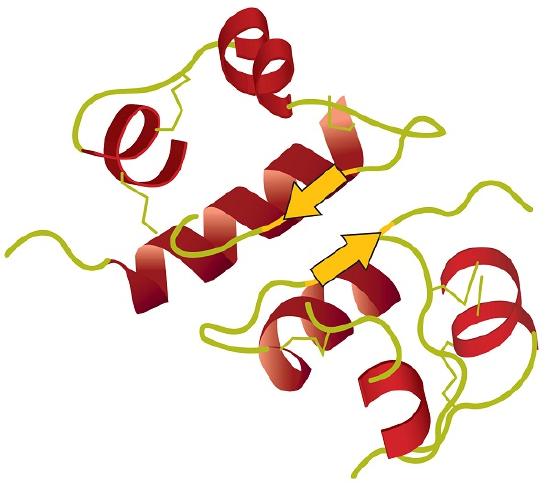
The general term peptide refers to an amino acid chain of unspecified length. However, chains of about 50 amino acids or more are usually called proteins or polypeptides. In its physiologically active form, a protein may be composed of one or more polypeptide chains. As an example, consider the 3-dimensional shape of the protein bradykinin, a protein important for vasodilation in the body (Figure 28.3j.):
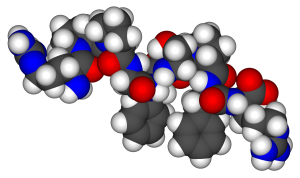
For peptides and proteins to be physiologically active, it is not enough that they incorporate certain amounts of specific amino acids. The order, or sequence, in which the amino acids are connected is also of critical importance. Bradykinin is a nine-amino acid peptide (Figure 28.3j.) produced in the blood that has the following amino acid sequence:
arg-pro-pro-gly-phe-ser-pro-phe-arg
This peptide lowers blood pressure, stimulates smooth muscle tissue, increases capillary permeability, and causes pain. When the order of amino acids in bradykinin is reversed,
arg-phe-pro-ser-phe-gly-pro-pro-arg
the peptide resulting from this synthesis shows none of the activity of bradykinin.
Just as millions of different words are spelled with our 26-letter English alphabet, millions of different proteins are made with the 20 common amino acids. However, just as the English alphabet can be used to write gibberish, amino acids can be put together in the wrong sequence to produce nonfunctional proteins. Although the correct sequence is ordinarily of utmost importance, it is not always absolutely required. Just as you can sometimes make sense of incorrectly spelled English words, a protein with a small percentage of “incorrect” amino acids may continue to function. However, it rarely functions as well as a protein having the correct sequence. There are also instances in which seemingly minor errors of sequence have disastrous effects. For example, in some people, every molecule of hemoglobin (a protein in the blood that transports oxygen) has a single incorrect amino acid unit out of about 300 (a single valine replaces a glutamic acid). That “minor” error is responsible for sickle cell anemia, an inherited condition that usually is fatal.
Proteins
Each of the thousands of naturally occurring proteins has its own characteristic amino acid composition and sequence that result in a unique three-dimensional shape. Since the 1950s, scientists have determined the amino acid sequences and three-dimensional conformation of numerous proteins and thus obtained important clues on how each protein performs its specific function in the body.
Proteins are compounds of high molar mass consisting largely or entirely of chains of amino acids. Because of their great complexity, protein molecules cannot be classified on the basis of specific structural similarities, as carbohydrates and lipids are categorized. The two major structural classifications of proteins are based on far more general qualities: whether the protein is (1) fiberlike and insoluble or (2) globular and soluble. Some proteins, such as those that compose hair, skin, muscles, and connective tissue, are fiberlike. These fibrous proteins are insoluble in water and usually serve structural, connective, and protective functions. Examples of fibrous proteins are keratins, collagens, myosins, and elastins. Hair and the outer layer of skin are composed of keratin. Connective tissues contain collagen. Myosins are muscle proteins and are capable of contraction and extension. Elastins are found in ligaments and the elastic tissue of artery walls.
Globular proteins, the other major class, are soluble in aqueous media. In these proteins, the chains are folded so that the molecule as a whole is roughly spherical. Familiar examples include egg albumin from egg whites and serum albumin in blood. Serum albumin plays a major role in transporting fatty acids and maintaining a proper balance of osmotic pressures in the body. Hemoglobin and myoglobin, which are important for binding oxygen, are also globular proteins.
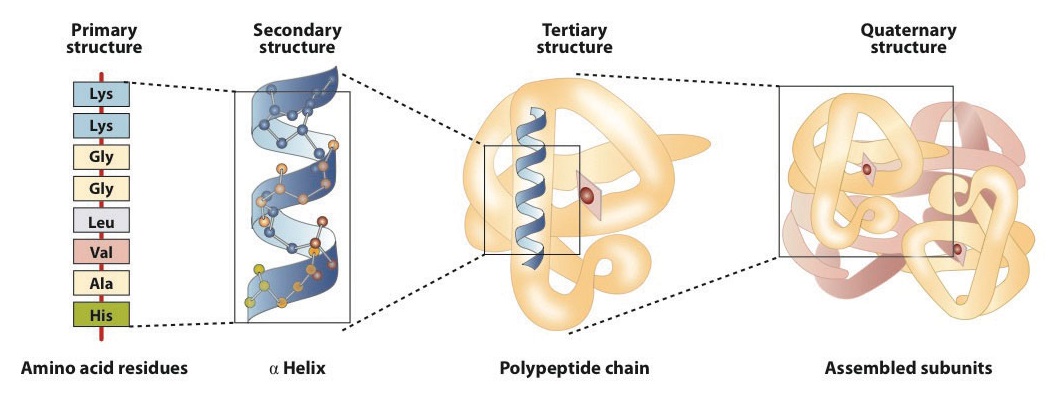
Levels of Protein Structure
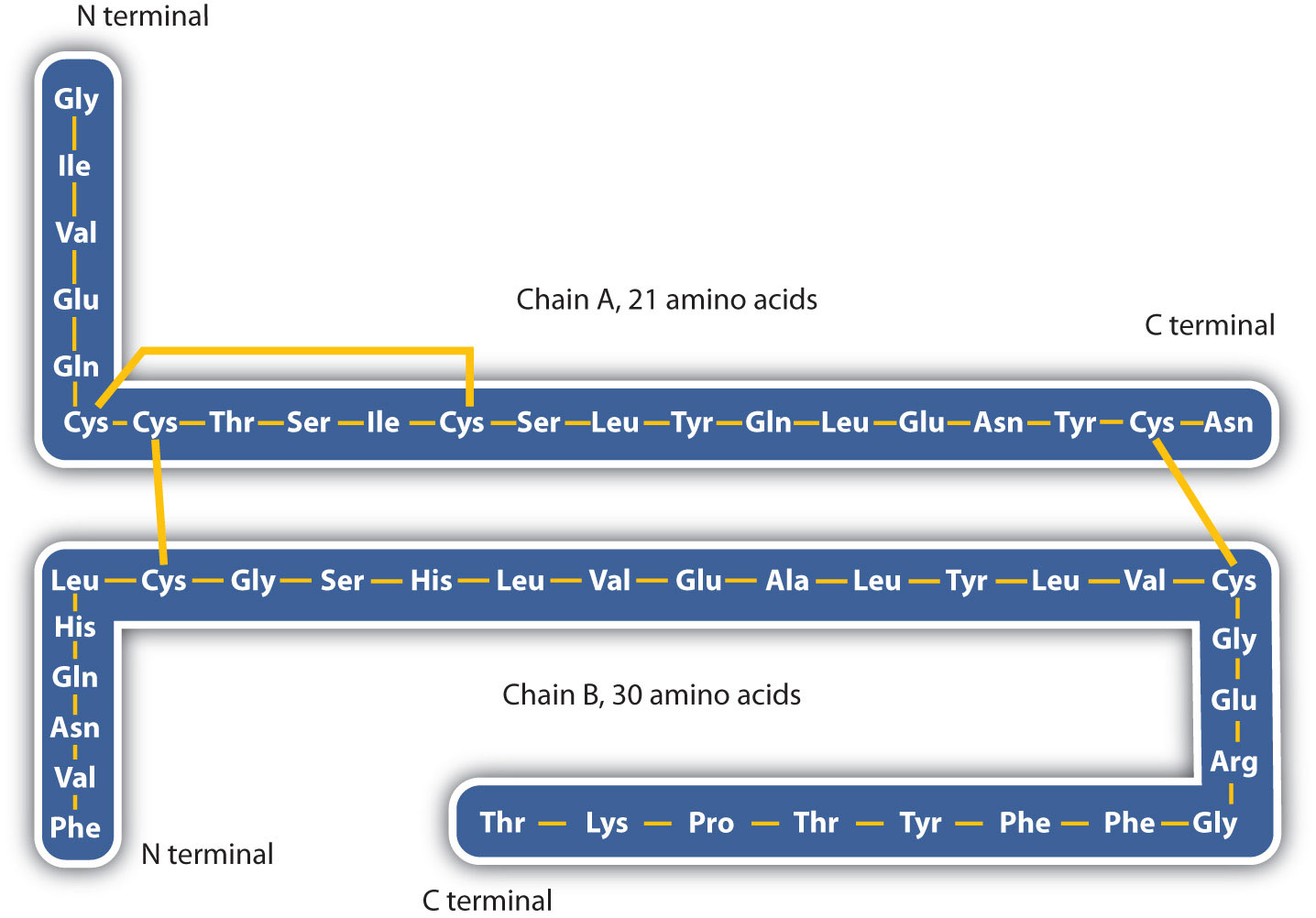
The structure of proteins is generally described as having four organizational levels. The first of these is the primary structure, which is the number and sequence of amino acids in a protein’s polypeptide chain or chains, beginning with the free amino group and maintained by the peptide bonds connecting each amino acid to the next. The primary structure of insulin, composed of 51 amino acids, is shown in figure 283k.
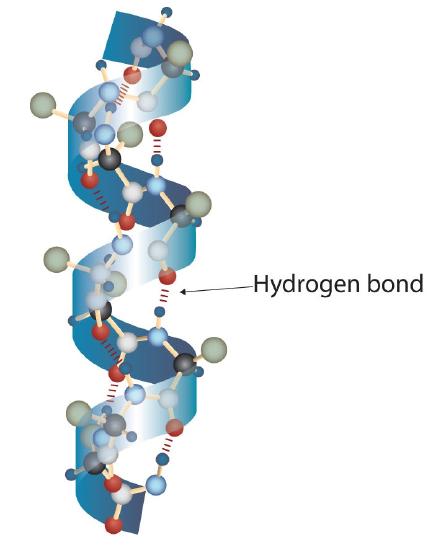
A protein molecule is not a random tangle of polypeptide chains. Instead, the chains are arranged in unique but specific conformations. The term secondary structure refers to the fixed arrangement of the polypeptide backbone. On the basis of X ray studies, Linus Pauling and Robert Corey postulated that certain proteins or portions of proteins twist into a spiral or a helix. This helix is stabilized by intrachain hydrogen bonding between the carbonyl oxygen atom of one amino acid and the amide hydrogen atom four amino acids up the chain (located on the next turn of the helix) and is known as a right-handed α-helix. X ray data indicate that this helix makes one turn for every 3.6 amino acids, and the side chains of these amino acids project outward from the coiled backbone (Figure 28.3l.). The α-keratins, found in hair and wool, are exclusively α-helical in conformation. Some proteins, such as gamma globulin, chymotrypsin, and cytochrome c, have little or no helical structure. Others, such as hemoglobin and myoglobin, are helical in certain regions but not in others.
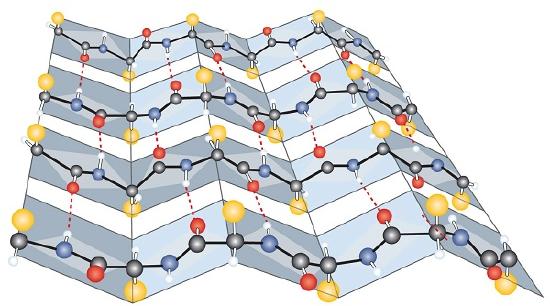
Another common type of secondary structure, called the β-pleated sheet conformation, is a sheetlike arrangement in which two or more extended polypeptide chains (or separate regions on the same chain) are aligned side by side. The aligned segments can run either parallel or antiparallel—that is, the N-terminals can face in the same direction on adjacent chains or in different directions—and are connected by interchain hydrogen bonding (Figure 28.3m.). The β-pleated sheet is particularly important in structural proteins, such as silk fibroin. It is also seen in portions of many enzymes, such as carboxypeptidase A and lysozyme.
Tertiary structure refers to the unique three-dimensional shape of the protein as a whole, which results from the folding and bending of the protein backbone. The tertiary structure is intimately tied to the proper biochemical functioning of the protein. Figure 28.3n. shows a depiction of the three-dimensional structure of insulin.
Four major types of attractive interactions determine the shape and stability of the tertiary structure of proteins. These are shown in Figure 28.3o.
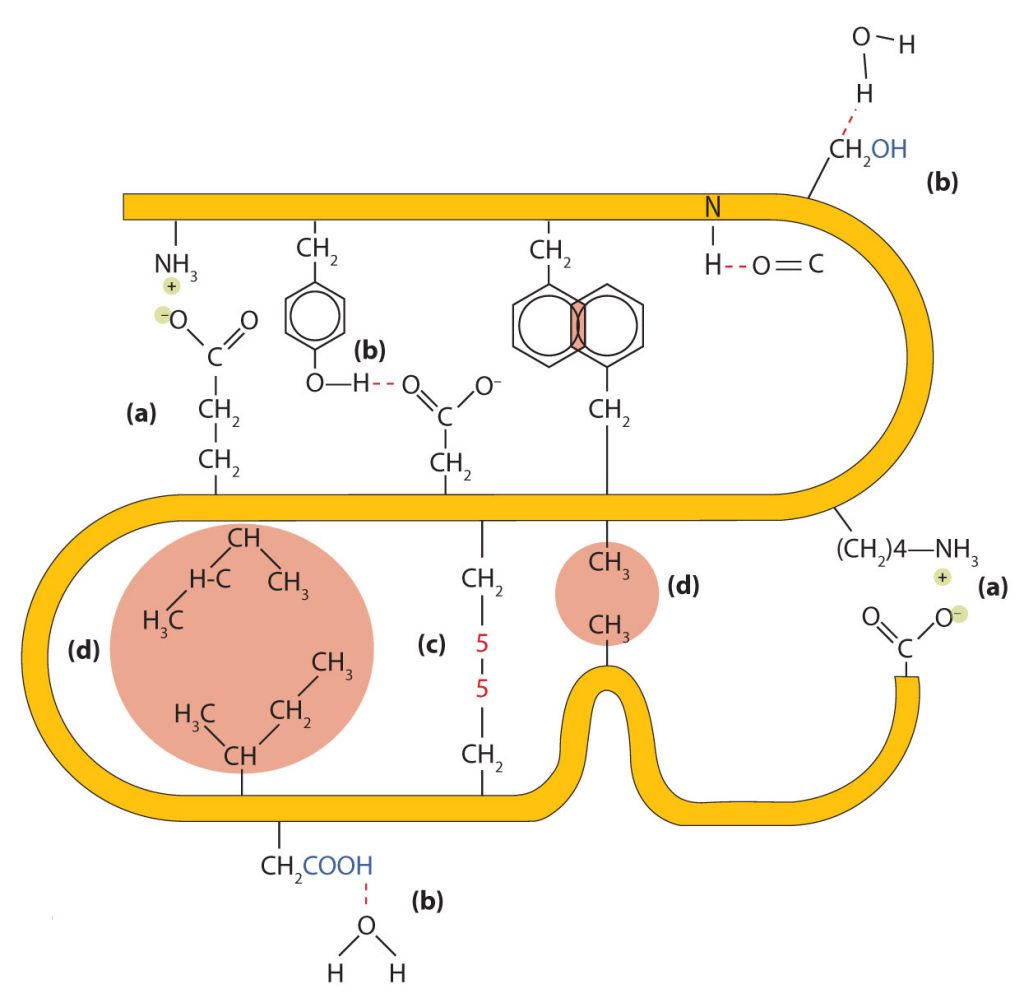
- Ionic bonding. Ionic bonds result from electrostatic attractions between positively and negatively charged side chains of amino acids. For example, the mutual attraction between an aspartic acid carboxylate ion and a lysine ammonium ion helps to maintain a particular folded area of a protein (part (a) of Figure 28.3o.).
- Hydrogen bonding. Hydrogen bonding forms between a highly electronegative oxygen atom or a nitrogen atom and a hydrogen atom attached to another oxygen atom or a nitrogen atom, such as those found in polar amino acid side chains. Hydrogen bonding (as well as ionic attractions) is extremely important in both the intra- and intermolecular interactions of proteins (part (b) of Figure 28.3o.).
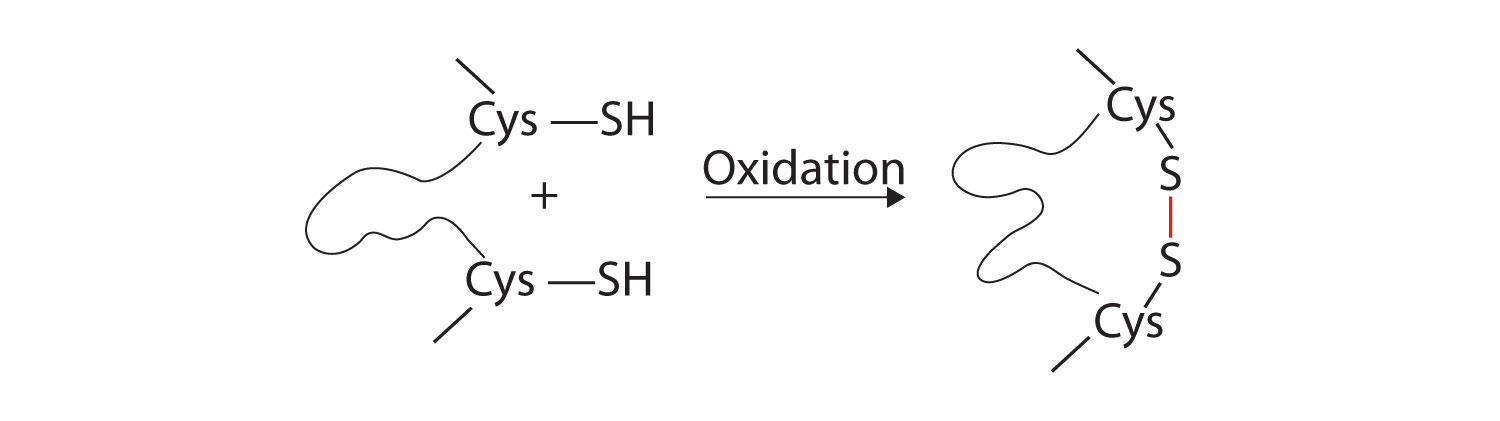
- Disulfide linkages. Two cysteine amino acid units may be brought close together as the protein molecule folds. Subsequent oxidation and linkage of the sulfur atoms in the highly reactive sulfhydryl (SH) groups leads to the formation of cystine (part (c) of Figure 28.3o. and figure 28.3p.). Intrachain disulfide linkages are found in many proteins, including insulin (yellow bars in Figure 28.3k.) and have a strong stabilizing effect on the tertiary structure.
- Dispersion forces. Dispersion forces arise when a normally nonpolar atom becomes momentarily polar due to an uneven distribution of electrons, leading to an instantaneous dipole that induces a shift of electrons in a neighbouring nonpolar atom. Dispersion forces are weak but can be important when other types of interactions are either missing or minimal (part (d) of Figure 28.3o.). This is the case with fibroin, the major protein in silk, in which a high proportion of amino acids in the protein have nonpolar side chains. The term hydrophobic interaction is often misused as a synonym for dispersion forces. Hydrophobic interactions arise because water molecules engage in hydrogen bonding with other water molecules (or groups in proteins capable of hydrogen bonding). Because nonpolar groups cannot engage in hydrogen bonding, the protein folds in such a way that these groups are buried in the interior part of the protein structure, minimizing their contact with water.
When a protein contains more than one polypeptide chain, each chain is called a subunit. The arrangement of multiple subunits represents a fourth level of structure, the quaternary structure of a protein. Hemoglobin, with four polypeptide chains or subunits, is the most frequently cited example of a protein having quaternary structure (Figure 28.3q.). The quaternary structure of a protein is produced and stabilized by the same kinds of interactions that produce and maintain the tertiary structure.
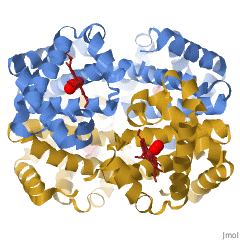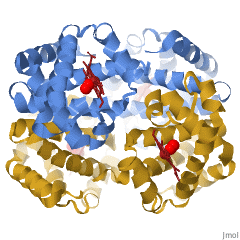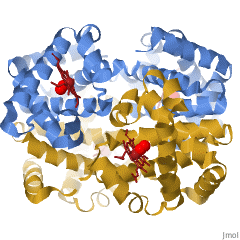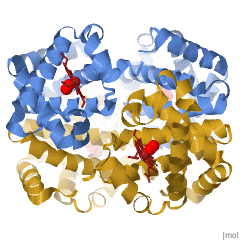
A schematic representation of the four levels of protein structure is shown in Figure 28.3r. The primary structure consists of the specific amino acid sequence. The resulting peptide chain can twist into an α-helix, which is one type of secondary structure. This helical segment is incorporated into the tertiary structure of the folded polypeptide chain. The single polypeptide chain is a subunit that constitutes the quaternary structure of a protein, such as hemoglobin that has four polypeptide chains.
Denaturation of Proteins
The highly organized structures of proteins are truly masterworks of chemical architecture. But highly organized structures tend to have a certain delicacy, and this is true of proteins. Denaturation is the term used for any change in the three-dimensional structure of a protein that renders it incapable of performing its assigned function. A denatured protein cannot do its job. (Sometimes denaturation is equated with the precipitation or coagulation of a protein; our definition is a bit broader.) A wide variety of reagents and conditions, such as heat, organic compounds, pH changes, and heavy metal ions can cause protein denaturation (Table 28.3f.).
| Method | Effect on Protein Structure |
|---|---|
| Heat above 50°C or ultraviolet (UV) radiation | Heat or UV radiation supplies kinetic energy to protein molecules, causing their atoms to vibrate more rapidly and disrupting relatively weak hydrogen bonding and dispersion forces. |
| Use of organic compounds, such as ethyl alcohol | These compounds are capable of engaging in intermolecular hydrogen bonding with protein molecules, disrupting intramolecular hydrogen bonding within the protein. |
| Salts of heavy metal ions, such as mercury, silver, and lead | These ions form strong bonds with the carboxylate anions of the acidic amino acids or SH groups of cysteine, disrupting ionic bonds and disulfide linkages. |
| Alkaloid reagents, such as tannic acid (used in tanning leather) | These reagents combine with positively charged amino groups in proteins to disrupt ionic bonds. |
Source: “18.4: Proteins” In Basics of GOB Chemistry (Ball et al.), CC BY-NC-SA 4.0.
Anyone who has fried an egg has observed denaturation. The clear egg white turns opaque as the albumin denatures and coagulates. No one has yet reversed that process. However, given the proper circumstances and enough time, a protein that has unfolded under sufficiently gentle conditions can refold and may again exhibit biological activity (Figure 28.3s.). Such evidence suggests that, at least for these proteins, the primary structure determines the secondary and tertiary structure. A given sequence of amino acids seems to adopt its particular three-dimensional arrangement naturally if conditions are right.
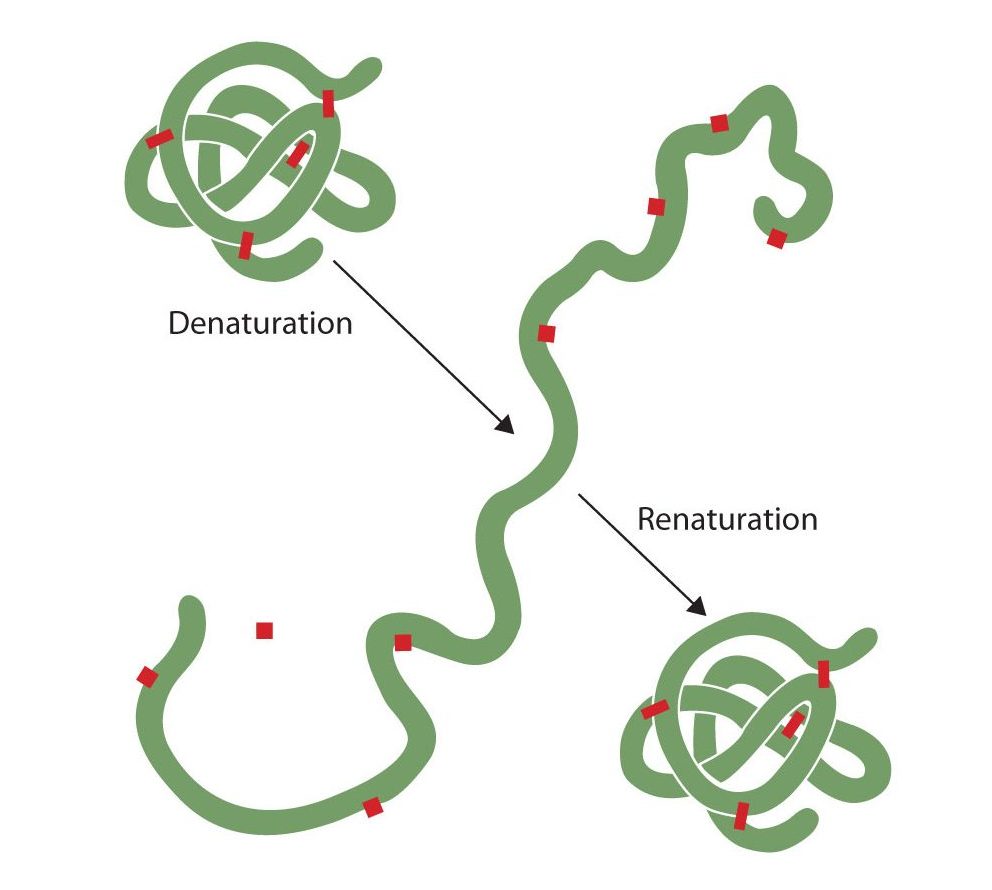
The primary structures of proteins are quite sturdy. In general, fairly vigorous conditions are needed to hydrolyze peptide bonds. At the secondary through quaternary levels, however, proteins are quite vulnerable to attack, though they vary in their vulnerability to denaturation. The delicately folded globular proteins are much easier to denature than are the tough, fibrous proteins of hair and skin.
Enzymes
A catalyst is any substance that increases the rate or speed of a chemical reaction without being changed or consumed in the reaction. Enzymes are biological catalysts, and nearly all of them are proteins. The reaction rates attained by enzymes are truly amazing. In their presence, reactions occur at rates that are a million (106) or more times faster than would be attainable in their absence. What is even more amazing is that enzymes perform this function at body temperature (~37°C) and physiological pH (pH ~7), rather than at the conditions that are typically necessary to increase reaction rates (high temperature or pressure, the use of strong oxidizing or reducing agents or strong acids or bases, or a combination of any of these). In addition, enzymes are highly specific in their action; that is, each enzyme catalyzes only one type of reaction in only one compound or a group of structurally related compounds. The compound or compounds on which an enzyme acts are known as its substrates.
Hundreds of enzymes have been purified and studied in an effort to understand how they work so effectively and with such specificity. The resulting knowledge has been used to design drugs that inhibit or activate particular enzymes. An example is the intensive research to improve the treatment of or find a cure for acquired immunodeficiency syndrome (AIDS). AIDS is caused by the human immunodeficiency virus (HIV). Researchers are studying the enzymes produced by this virus and are developing drugs intended to block the action of those enzymes without interfering with enzymes produced by the human body. Several of these drugs have now been approved for use by AIDS patients.
The first enzymes to be discovered were named according to their source or method of discovery. The enzyme pepsin, which aids in the hydrolysis of proteins, is found in the digestive juices of the stomach (Greek pepsis, meaning “digestion”). Papain, another enzyme that hydrolyzes protein (in fact, it is used in meat tenderizers), is isolated from papayas. As more enzymes were discovered, chemists recognized the need for a more systematic and chemically informative identification scheme. In the current numbering and naming scheme, under the oversight of the Nomenclature Commission of the International Union of Biochemistry, enzymes are arranged into six groups according to the general type of reaction they catalyze (Table 28.3g.), with subgroups and secondary subgroups that specify the reaction more precisely.
| Class | Type of Reaction Catalyzed | Examples |
|---|---|---|
| oxidoreductases | oxidation-reduction reactions | Dehydrogenases catalyze oxidation-reduction reactions involving hydrogen and reductases catalyze reactions in which a substrate is reduced. |
| transferases | transfer reactions of groups, such as methyl, amino, and acetyl | Transaminases catalyze the transfer of amino group, and kinases catalyze the transfer of a phosphate group. |
| hydrolases | hydrolysis reactions | Lipases catalyze the hydrolysis of lipids, and proteases catalyze the hydrolysis of proteins |
| lyases | reactions in which groups are removed without hydrolysis or addition of groups to a double bond | Decarboxylases catalyze the removal of carboxyl groups. |
| isomerases | reactions in which a compound is converted to its isomer | Isomerases may catalyze the conversion of an aldose to a ketose, and mutases catalyze reactions in which a functional group is transferred from one atom in a substrate to another. |
| ligases | reactions in which new bonds are formed between carbon and another atom; energy is required | Synthetases catalyze reactions in which two smaller molecules are linked to form a larger one. |
Source: “18.5: Enzymes” In Basics of GOB Chemistry (Ball et al.), CC BY-NC-SA 4.0.
Each enzyme is assigned a four-digit number, preceded by the prefix EC—for enzyme classification—that indicates its group, subgroup, and so forth. This is demonstrated in Table 28.19b. for alcohol dehydrogenase. Each enzyme is also given a name consisting of the root of the name of its substrate or substrates and the –ase suffix. Thus urease is the enzyme that catalyzes the hydrolysis of urea.
Example 28.3b
Alcohol dehydrogenase has an Enzyme Classification Number of EC 1.1.1.1. What does this mean?
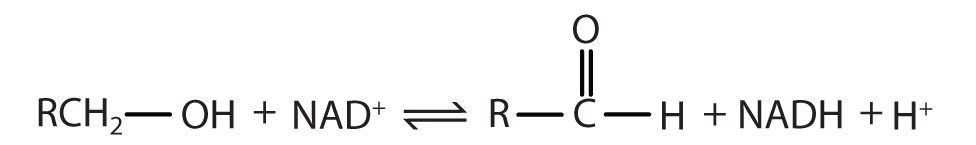
Solution:
The first digit indicates that this enzyme is an oxidoreductase; that is, an enzyme that catalyzes an oxidation-reduction reaction. The second digit indicates that this oxidoreductase catalyzes a reaction involving a primary or secondary alcohol. The third digit indicates that either the coenzyme NAD+ or NADP+ is required for this reaction. The fourth digit indicates that this was the first enzyme isolated, characterized, and named using this system of nomenclature. The systematic name for this enzyme is alcohol:NAD+ oxidoreductase, while the recommended or common name is alcohol dehydrogenase.
Enzyme Action
Enzyme-catalyzed reactions occur in at least two steps. In the first step, an enzyme molecule (E) and the substrate molecule or molecules (S) collide and react to form an intermediate compound called the enzyme-substrate (E–S) complex. (This step is reversible because the complex can break apart into the original substrate or substrates and the free enzyme.) Once the E–S complex forms, the enzyme is able to catalyze the formation of product (P), which is then released from the enzyme surface:
[latex]S + E \rightarrow E–S \tag{1}[/latex]
[latex]E–S \rightarrow P + E \tag{2}[/latex]
Hydrogen bonding and other electrostatic interactions hold the enzyme and substrate together in the complex. The structural features or functional groups on the enzyme that participate in these interactions are located in a cleft or pocket on the enzyme surface. This pocket, where the enzyme combines with the substrate and transforms the substrate to product is called the active site of the enzyme (Figure 28.3t.).
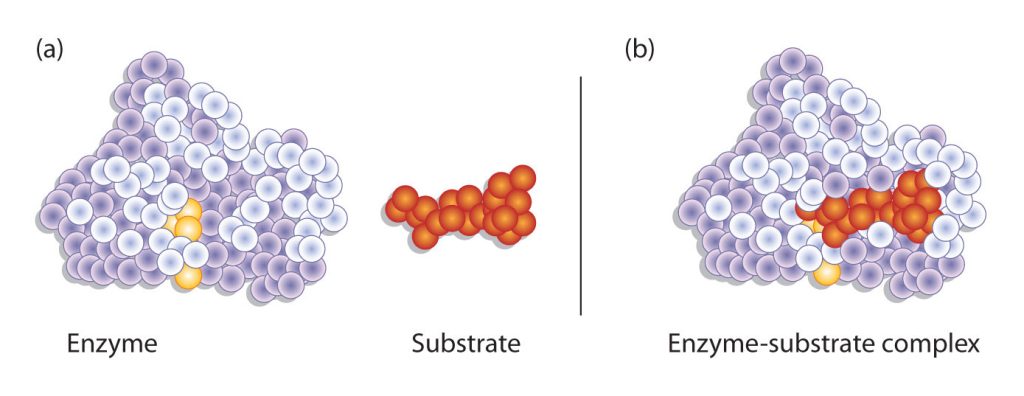
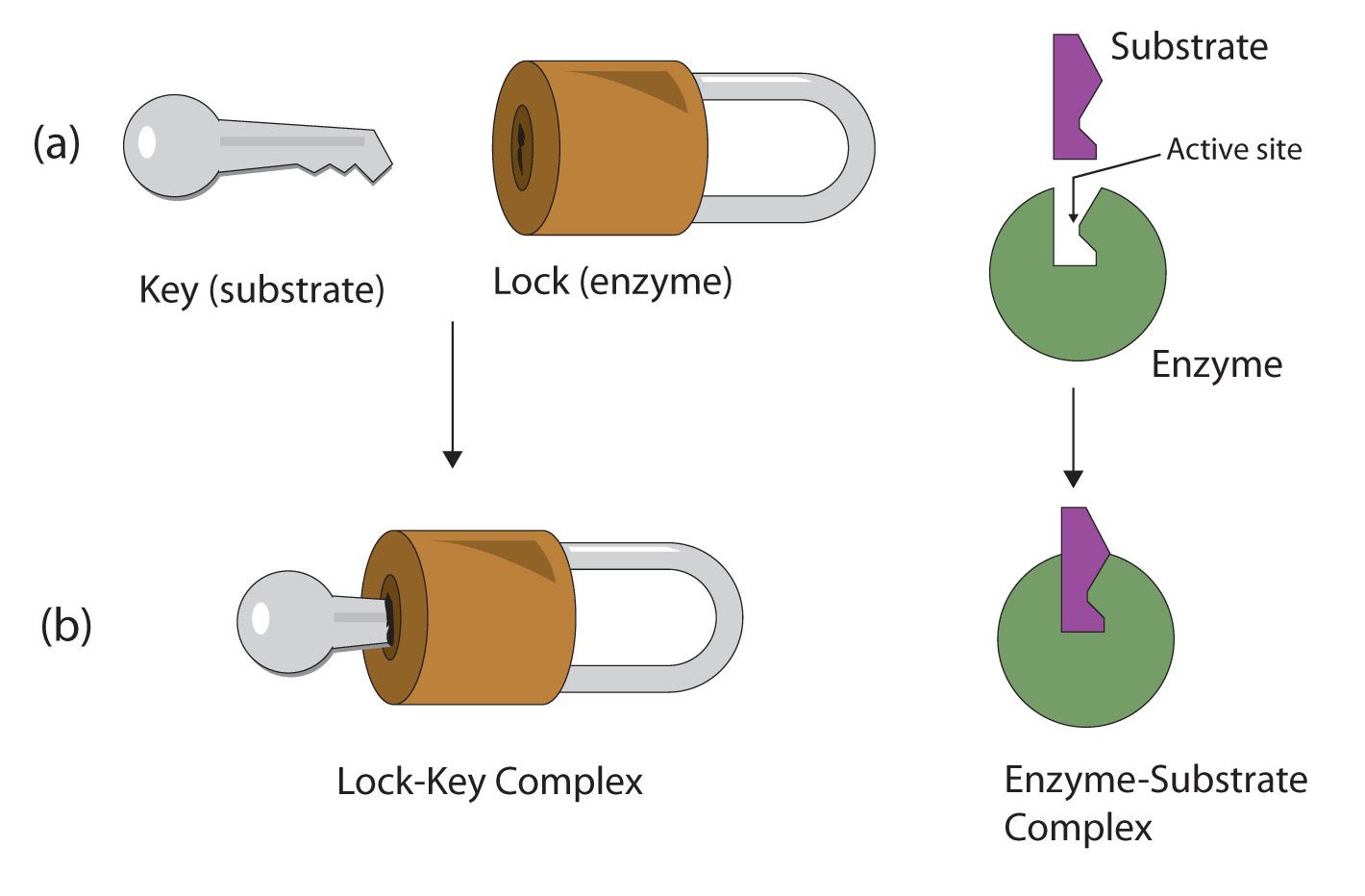
The active site of an enzyme possesses a unique conformation (including correctly positioned bonding groups) that is complementary to the structure of the substrate, so that the enzyme and substrate molecules fit together in much the same manner as a key fits into a tumbler lock. In fact, an early model describing the formation of the enzyme-substrate complex was called the lock-and-key model (Figure 28.3u.). This model portrayed the enzyme as conformationally rigid and able to bond only to substrates that exactly fit the active site.
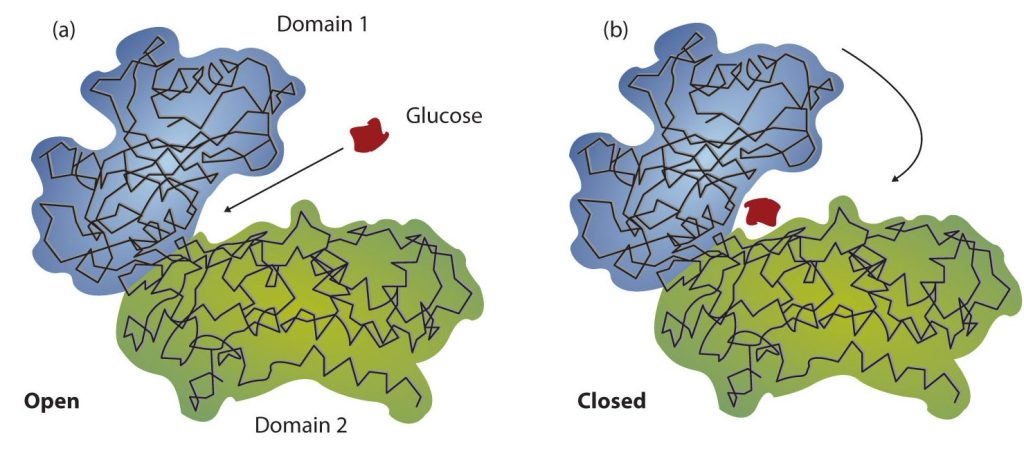
Working out the precise three-dimensional structures of numerous enzymes has enabled chemists to refine the original lock-and-key model of enzyme actions. They discovered that the binding of a substrate often leads to a large conformational change in the enzyme, as well as to changes in the structure of the substrate or substrates. The current theory, known as the induced-fit model, says that enzymes can undergo a change in conformation when they bind substrate molecules, and the active site has a shape complementary to that of the substrate only after the substrate is bound, as shown for hexokinase in Figure 28.3v. After catalysis, the enzyme resumes its original structure.
The structural changes that occur when an enzyme and a substrate join together bring specific parts of a substrate into alignment with specific parts of the enzyme’s active site. Amino acid side chains in or near the binding site can then act as acid or base catalysts, provide binding sites for the transfer of functional groups from one substrate to another or aid in the rearrangement of a substrate. The participating amino acids, which are usually widely separated in the primary sequence of the protein, are brought close together in the active site as a result of the folding and bending of the polypeptide chain or chains when the protein acquires its tertiary and quaternary structure. Binding to enzymes brings reactants close to each other and aligns them properly, which has the same effect as increasing the concentration of the reacting compounds.
- What type of interaction would occur between an OH group present on a substrate molecule and a functional group in the active site of an enzyme?
- Suggest an amino acid whose side chain might be in the active site of an enzyme and form the type of interaction you just identified.
Solution
- An OH group would most likely engage in hydrogen bonding with an appropriate functional group present in the active site of an enzyme.
- Several amino acid side chains would be able to engage in hydrogen bonding with an OH group. One example would be asparagine, which has an amide functional group.
One characteristic that distinguishes an enzyme from all other types of catalysts is its substrate specificity. An inorganic acid such as sulfuric acid can be used to increase the reaction rates of many different reactions, such as the hydrolysis of disaccharides, polysaccharides, lipids, and proteins, with complete impartiality. In contrast, enzymes are much more specific. Some enzymes act on a single substrate, while other enzymes act on any of a group of related molecules containing a similar functional group or chemical bond. Some enzymes even distinguish between D- and L-stereoisomers, binding one stereoisomer but not the other. Urease, for example, is an enzyme that catalyzes the hydrolysis of a single substrate—urea—but not the closely related compounds methyl urea, thiourea, or biuret (Figure 28.3w.). The enzyme carboxypeptidase, on the other hand, is far less specific. It catalyzes the removal of nearly any amino acid from the carboxyl end of any peptide or protein.
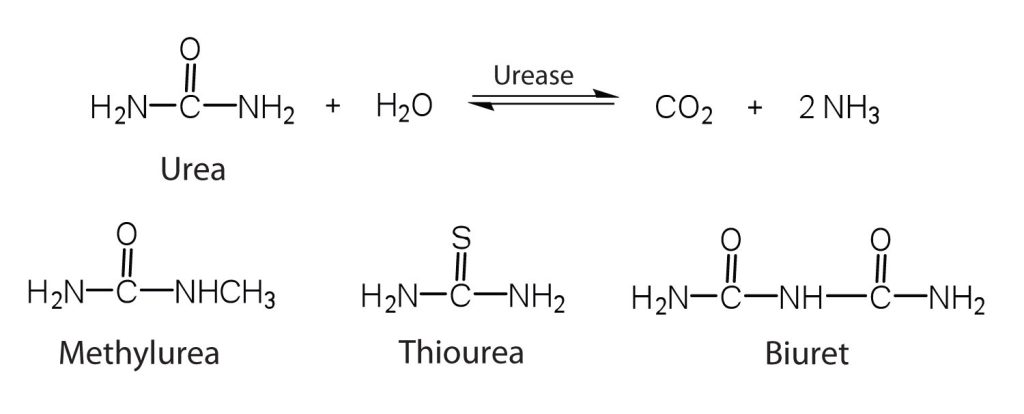
Enzyme specificity results from the uniqueness of the active site in each different enzyme because of the identity, charge, and spatial orientation of the functional groups located there. It regulates cell chemistry so that the proper reactions occur in the proper place at the proper time. Clearly, it is crucial to the proper functioning of the living cell.
Attribution & References
Except where otherwise noted, portions of this page were written by Gregory A. Anderson while others were adapted by Gregory A. Anderson and Samantha Sullivan Sauer from “18.0: Prelude to Amino Acids, Proteins, and Enzymes“, “18.1: Properties of Amino Acids“, “18.2: Reactions of Amino Acids“, “18.3: Peptides“, “18.4: Proteins“, “18.5: Enzymes“, & “18.6: Enzyme Action” Basics of General, Organic, and Biological Chemistry (Ball et al.) by David W. Ball, John W. Hill, and Rhonda J. Scott via LibreTexts, CC BY-NC-SA 4.0./ A LibreTexts version of Introduction to Chemistry: GOB (v. 1.0), CC BY-NC 3.0.
-
- glycine, alanine, valine, leucine, and isoleucine all have alkane based R groups which are non-polar due to the carbon-hydrogen components. Phenylalanine is aromatic and thus non-polar. Tryptophane and proline have amine and hydrocarbon R groups which are non-polar. Methionine has a S-C based R group and is non-polar as the electronegativity of S is the same as C.
- Serine, threonine and tyrosine have alcohol functional groups in their R groups. Oxygen is electronegative resulting in a polar region. Cysteine has a thiol group and S is electronegative compared to H. Asparagine and glutamine have carbonyl groups and the O makes the region polar.
↵

