9. Microbial Growth and Biosynthesis
9.1 Biosynthesis
Learning Objectives
- Describe the importance of substrates for biosynthesis
- Identify the roles of the central pathways in biosynthesis
- Describe the various ways autotrophic microorganisms fix carbon
- Describe the importance of nitrogen fixation
- Explain the importance of assimilatory reduction of sulphur
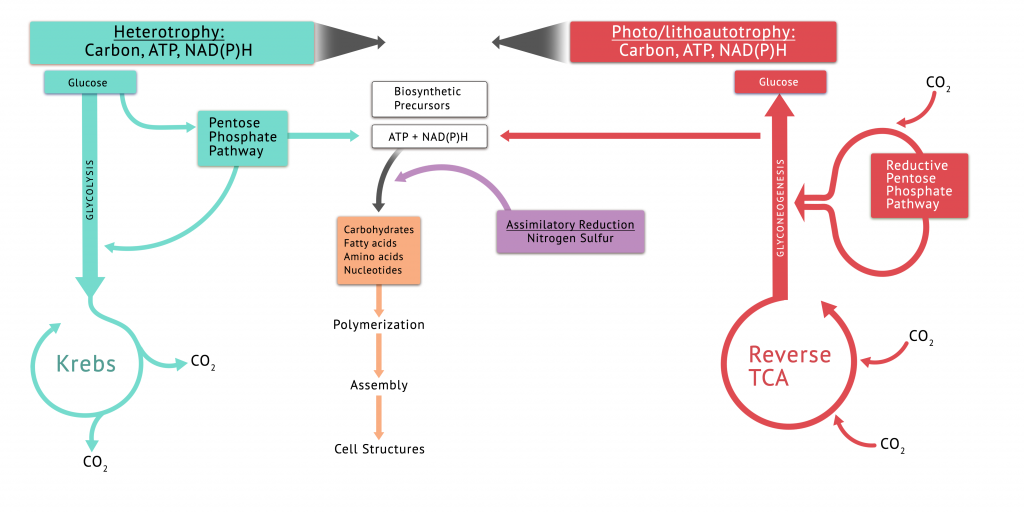
Cellular metabolism includes catabolic pathways, which extracts energy from the environment, and anabolic pathways, which use energy and carbon building blocks for cellular growth. Microbes obtain their energy through heterotrophy, lithotrophy (bacteria and archaea only) or phototrophy. Anabolic pathways are necessary for survival and cellular function, giving rise to larger, more complex molecules which store energy. They are, in contrast to microbial catabolism, relatively conserved among the microbes. So whether an organism is an autotroph (using CO2 as a carbon source) or heterotroph (using organic carbon sources), the anabolic pathways generally require the same small set of organic building blocks – the precursor metabolites. In heterotrophs, these molecules come from the reactions in their central catabolic pathways:
- Glycolytic pathway (= Embden Meyerhof Parnas pathway)
- Entner-Doudoroff pathway
- Pentose phosphate pathway
- TCA pathway (= tricarboxylic acid pathway, Kreb’s cycle or citric acid cycle)
Because many of the enzymatic reactions within these pathways are also used to create precursor molecules in autotrophs, the four pathways are called the central pathways and, in the heterotrophs, are referred to as amphibolic – they function in both the catabolic and anabolic directions.
Substrates for Biosynthesis
In heterotrophs, biosynthetic processes begin with diverting some of the intermediates in the central pathways, as well as the energy and reduced cofactors from those pathways. The balance between catabolic oxidation of organics, and the anabolic reduction of organics to build biomass, must therefore be tightly regulated.
TCA Cycle
The TCA cycle, also known as the citric acid cycle, or Krebs cycle, produces energy through the complete oxidation of acetate, derived from carbohydrates, fats, and proteins, to carbon dioxide. The cycle is one of the major metabolic processes utilized to generate energy, and it also provides precursors for a number of biosynthetic pathways. These precursors are used as substrates for the biogenesis of some of the amino acids, pyrimidines (nucleic acid bases), and reducing agents such as NADH.
Glycolytic pathway
The glycolytic pathway, also known as the Embden Meyerhof Parnas pathway) catalyzes a series of reactions that oxidize the 6 carbon sugar glucose, into the 3 carbon molecule pyruvate. Most of these reactions are reversible, and with the addition of a few enzymes that function only in the reductive direction, the pathway can also be reversed to synthesize glucose from pyruvate. This is called gluconeogenesis and is important for heterotrophs growing on organics other than glucose or its polymers, as well as for autotrophs (Figure 9.2). This pathway is depicted below. From glucose, other carbohydrates can be synthesized. In combination with the pentose phosphate pathway, precursor molecules are provided for biosynthesis of certain amino acids, nucleosides and purines (nucleic acid bases). The figure below shows the gluconeogenic pathway.
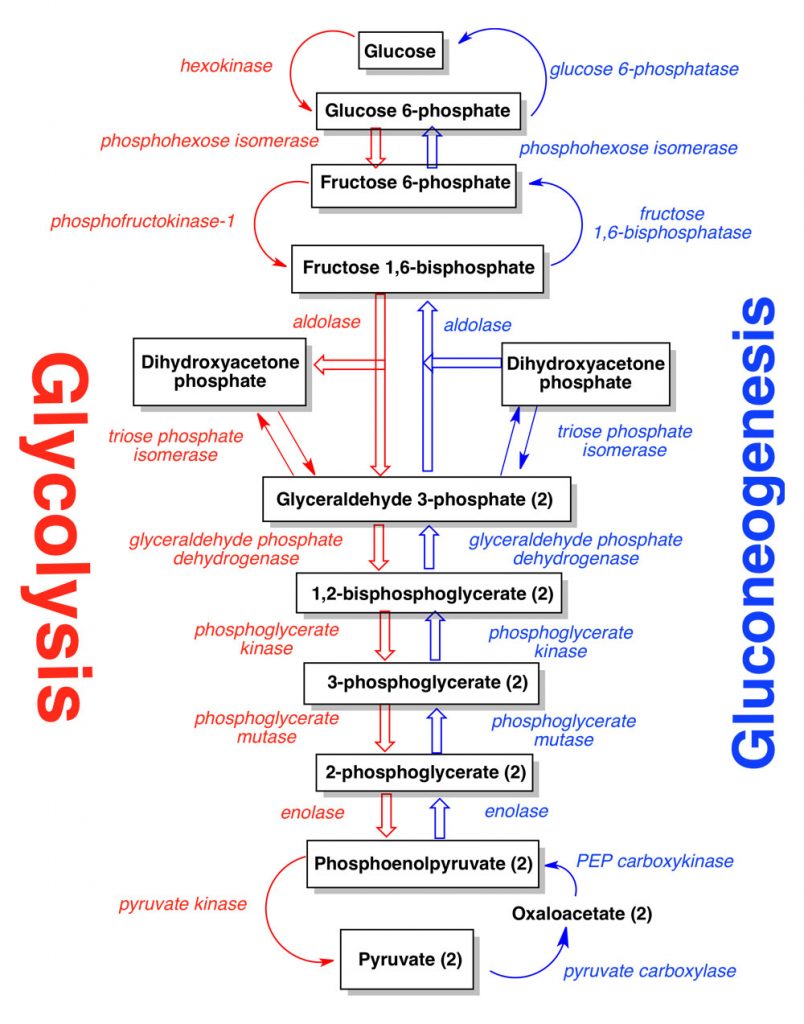
Many bacteria and archaea use an alternative to the glycolytic pathway called the Entner Doudoroff pathway. This pathway begins with glucose 6-phosphate, produces pyruvate and glyceraldehyde 3-phosphate and also allows for the catabolism of sugar acids, in addition to various sugars. It requires less ATP than the glycolytic pathway and is therefore thought to have evolved earlier than the glycolytic pathway. The pathway is prevalent in the enteric bacteria of the human colon (e.g. E. coli), enabling these bacteria to catabolize the mucous produced by the intestinal epithelium.
- What is anabolism and how does it relate to precursor metabolites?
- How do heterotrophs obtain precursor metabolites?
- What is the relevance of the Entner Doudoroff pathway?
Autotrophy
Autotrophs obtain their carbon from carbon dioxide, “fixing” the gas into organic molecules and biomass. In any ecosystem, they represent the producers, or base, of the associated food chain. Microorganisms can fix CO2 using a number of different pathways: two common ones are the Calvin cycle and the reductive TCA cycle. Both produce reduced carbon compounds from carbon dioxide and water and are therefore assimilatory reduction pathways. Carbon fixation occurs within the cytoplasm of photosynthetic and lithotrophic bacteria and archaea, and in the stroma of eukaryotic chloroplasts. In the photosynthetic organisms, CO2-fixation has historically been referred to as the light-independent reactions.
The Calvin Cycle
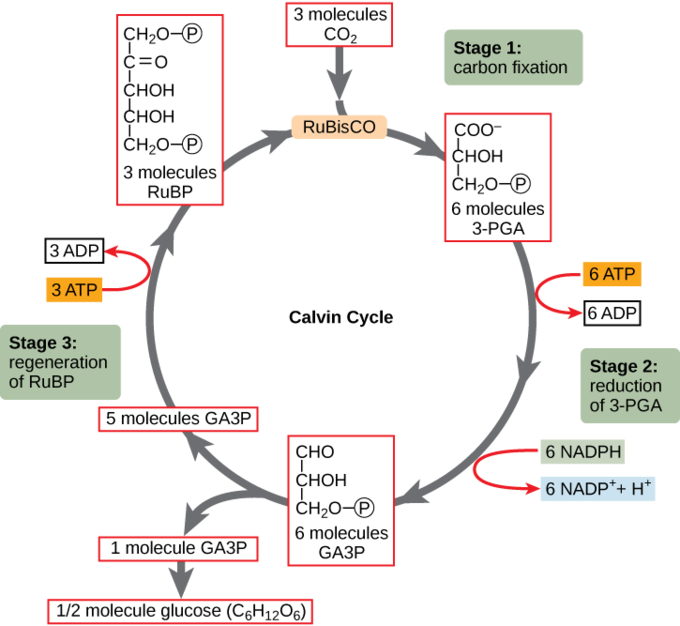
The Calvin cycle, also called the Calvin-Benson Cycle and the reductive pentose phosphate pathway, involves reducing carbon dioxide to the 3-carbon sugar phosphate, glyceraldehyde 3-phosphate (GA3P). Throughout the Calvin Cycle, intermediate molecules are consistently withdrawn to support growth. The cycle has three basic stages: fixation, reduction, and regeneration (Figure 9.3 & Appendix C).
- Fixation: The enzyme ribulose bisphosphate carboxylase (RuBisCO) catalyzes the addition of a CO2 to ribulose bisphosphate (RuBP). This results in the production of 3-phosphoglycerate (3-PGA).
- Reduction: Six molecules of both ATP and NADPH (from the light-dependent reactions) are used to convert 3-PGA into glyceraldehyde 3-phosphate (G3P). Some G3P is then used to build glucose.
- Regeneration: The remaining G3P not used to synthesize glucose is used to regenerate RuBP, enabling the system to continue CO2 fixation. Three more molecules of ATP are used in these regeneration reactions.
The Calvin cycle is used extensively by plants and photoautotrophic bacteria, and the enzyme RuBisCO is said to be the most plentiful enzyme on earth, comprising 30%–50% of the total soluble protein in plant chloroplasts.[1] However, besides its prevalent use in photoautotrophs, the Calvin cycle is also used by many lithotrophs to fix CO2. Some examples of microorganisms that utilize the Calvin cycle include cyanobacteria (whose ancestor gave rise to the eukaryotic chloroplast), purple bacteria, and nitrifying bacteria.
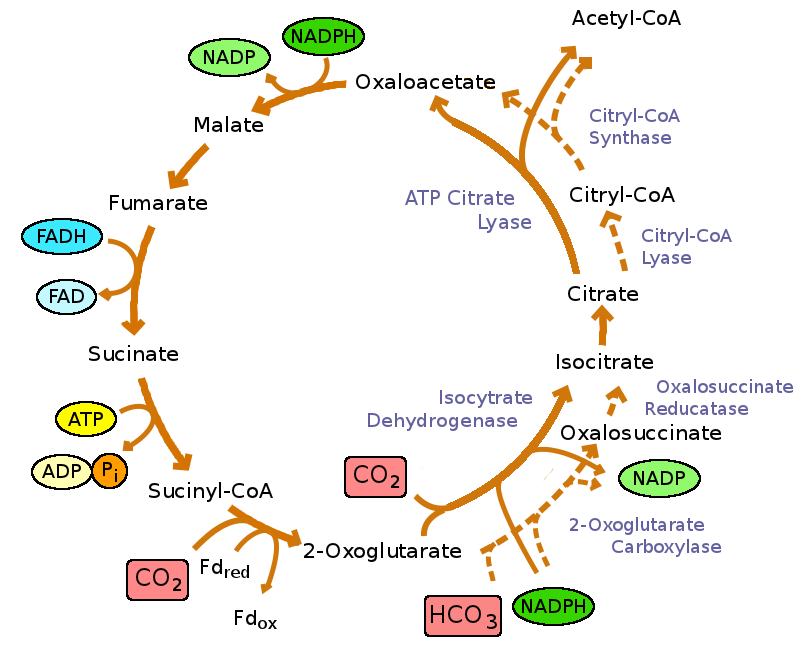
Reverse TCA cycle
This process, used by numerous anaerobic microorganisms, also produces organic compounds from carbon dioxide and water. The reactions are, as the name implies, the reverse of what is seen in the TCA cycle (Figure 9.4).
- What are the shared features of the Calvin cycle and the reverse TCA cycle?
- What is gluconeogenesis and how does the pathway relate to the glycolytic pathway?
- What is the reductive pentose phosphate pathway?
Assimilation of Nitrogen
Synthesis of the major nitrogen-containing compounds of the cell begins with precursor molecules from the central pathways. However none of the intermediates in the central pathways contain nitrogen. A source of nitrogen is a critical growth requirement, given its presence in the key cellular molecules:
- amino acids
- purines and pyrimidines
- porphyrins (required for numerous cellular structures including cytochromes and chlorophyll)
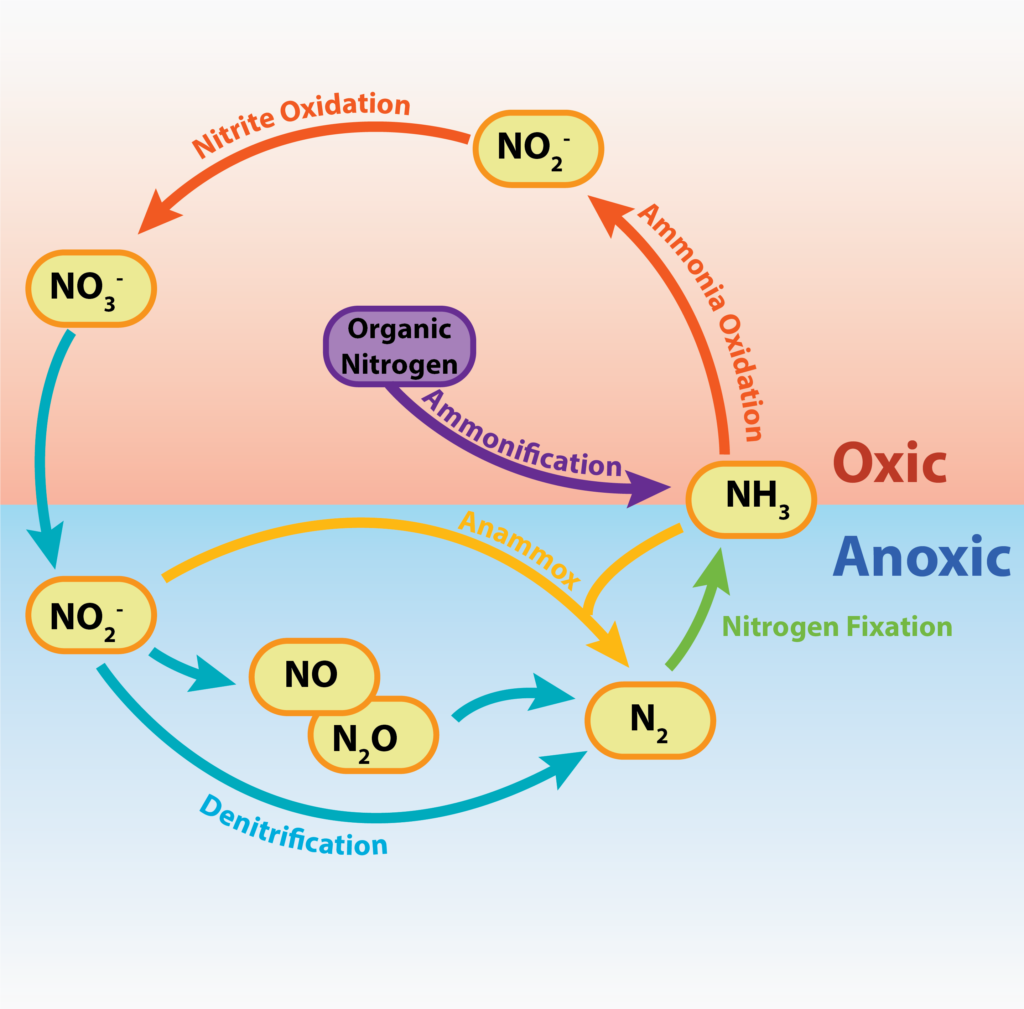
Different species can use different sources of nitrogen. However, unique to certain species of bacteria and archaea, is the ability to use atmospheric nitrogen, N2. Plants, animals, and other organisms rely on the nitrogen-fixers – the “diazotrophs” – since no eukaryote is known that can fix nitrogen, and only certain species of bacteria and archaea have the ability. The diazotrophs are therefore critically important to the global nitrogen cycle (Figure 9.5).
Nitrogen Fixation
Diazotrophy requires the unique enzyme nitrogenase, which converts N2 into ammonia (NH3), a much more useable form of nitrogen for most life forms. The process is extremely energetically costly, given the triple bond in N2 and the number of reduced cofactors required to reduce each atom of nitrogen to NH3. As a result, if a more easily metabolized nitrogen source such as NH3 or NO3– is available, diazotrophic organisms will use these instead of fixing N2. The nitrogenase enzyme is inactivated by O2, so the process also requires either an anaerobic environment or a mechanism by which the enzyme is protected from exposure to O2. Aerobic nitrogen-fixing organisms have evolved a variety of anaerobiosis strategies. Given the complexities of N-fixation, expression of the nitrogen-fixing genes, as well as the activity of the enzyme, is subject to stringent regulation.
Nitrogen-fixing organisms can either exist independently or symbiotically with a plant host:
- Symbiotic nitrogen-fixing organisms: these bacteria partner with a plant, to provide them with an environment appropriate for the functioning of their nitrogenase enzyme. The bacteria live in the plant’s tissue, fixing nitrogen and sharing the results (Figure 9.6). The plant provides both the location to fix nitrogen, the oxygen-binding protein leghaemoglobin, as well as additional nutrients to support the energy-taxing process of nitrogen fixation. The bacteria and the host exchange chemical recognition signals that facilitate the relationship. One of the best known bacteria in this category is Rhizobium, which partners with plants of the legume family (clover, soybeans, alfalfa, etc).
- Free-living nitrogen-fixing organisms: these organisms fix nitrogen for their own use but provide fixed nitrogen to other community members when it dies or is ingested. Free-living nitrogen-fixing organisms that grow anaerobically do not have to worry about special adaptations for their nitrogenase enzyme. Aerobic organisms must, however, establish anaerobiosis. Filamentous cyanobacteria make specialized cells known as heterocysts in which nitrogen fixation occurs (Figure 9.7). Since cyanobacteria produce oxygen as part of their photosynthesis, an anoxygenic version occurs within the heterocyst, allowing the nitrogenase to remain active. The heterocysts share the fixed nitrogen with surrounding cells, while the surrounding cells provide additional nutrients to the heterocysts. Some unicellular cyanobacteria use temporal separation of photosynthesis and diazotrophy, only fixing nitrogen in the dark. Other aerobic diazotrophs such as Azotobacter, use proteins that protect the nitrogenase from the effects of O2.

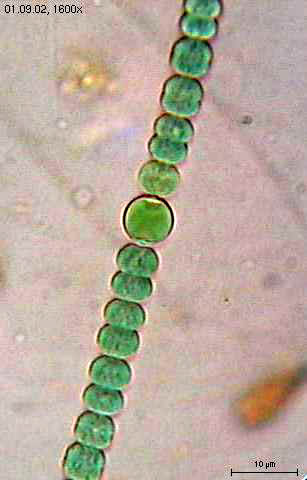
Assimilation into Organic Molecules
Inorganic forms of nitrogen are reduced to organic nitrogen compounds such as amino acids and nucleotides, allowing for cellular growth and reproduction. These reduction reactions or pathways are therefore assimilatory reductions. Ammonia assimilation occurs when the ammonia (NH3)/ammonium ion (NH4+) formed during nitrogen fixation is incorporated into cellular nitrogen. Assimilatory nitrate reduction (NO3–) to cellular nitrogen requires a multi-step process where nitrate is reduced to nitrite then ammonia and finally into organic nitrogen.
- What are diazotrophs and with which types of microbes is it associated?
- Why would diazotrophs prefer the nutrients NO3– and NH4+?
- What are the key features of the nitrogenase enzyme?
- What are some of the essential adaptations of aerobic diazotrophs?
Assimilation of Sulphur
Sulphur is another element that is present in major cellular molecules – for example the amino acids methionine and cysteine. Like nitrogen, it is absent from the precursors in the central pathways, and is incorporated through assimilatory processes. This is, however, a process seen in all organisms, although they may use different sources of inorganic sulphur nutrients. By far the most common source is SO42-, however certain bacteria and archaea can use alternative sources such as elemental sulphur (S0) or hydrogen sulphide (H2S). The latter is toxic to most other life forms.
Figure 9.8 provides a summary of biosynthesis in the heterotrophs and autotrophs, including the roles of the central pathways, and assimilatory reduction of sulphur and nitrogen.
Key Takeaways
- Anabolism is the form of metabolism responsible for building large complexes from precursors.
- The glycolytic and TCA central metabolic pathways produce precursors and substrates used in biosynthetic processes.
- The two major pathways for carbon fixation in the microbes are the Calvin, and reverse TCA cycles.
- Gluconeogenesis is a biosynthetic pathway that produces glucose from pyruvate.
- The reverse TCA cycle utilizes carbon dioxide and water to produce acetyl-CoA.
- The Calvin cycle reduces CO2 by adding it to the 5-carbon molecule ribulose bisphosphate to make 2 molecules of the 3-carbon gluconeogenesis intermediate, glyceraldehyde 3-phosphate .
- Diazotrophy, or nitrogen fixation, converts elemental nitrogen (N2) into ammonia, a reaction catalyzed by certain species of bacteria and archaea. The ammonia is immediately incorporated into amino air and other biosynthetic pathways for N-containing organics.
- Fixed nitrogen is an essential component of nucleic acids and proteins, therefore diazotrophic bacteria and archaea are essential to the global nitrogen cycle.
- Nitrogen fixation is catalyzed by the nitrogenase enzyme, and enzyme that is extremely sensitive to inactivation by exposure to oxygen.
- To protect nitrogenase from inactivation by oxygen, aerobic diazotrophs use physical or temporal separation, or oxygen-binding proteins, to protect nitrogenase from exposure to O2.
- Assimilatory reduction of sulphur nutrients is a universal ability
- Sulfate (SO42-) is the most common sulphur nutrient however certain bacteria and archaea can use sources such as S0 and H2S.
Multiple Choice
Short Answer
- What are the central pathways of the heterotrophs and why are they referred to as “amphibolic”?
- Which elements are missing from the central pathways but present in various major cellular polymers?
Critical Thinking
- Given that autotrophs acquire their energy from sunlight or inorganic compounds, why would they have the majority of enzymes from the central pathways of the heterotrophs?
- Compare the role of gluconeogenesis in heterotrophs with its role in autotrophs.
Media Attributions
- Glycolytic_and_gluconeogenic_pathways
- Calvin cycle
- Reductive_TCA_cycle
- Nitrogen-Cycle-1024×1009
- Rhizobia_nodules_on_Vigna_unguiculata
- Anabaena_sperica2
- biosynthesis-chart-ver-3.5
- A. Dhingra et al. “Enhanced Translation of a Chloroplast-Expressed RbcS Gene Restores Small Subunit Levels and Photosynthesis in Nuclear RbcS Antisense Plants.” Proceedings of the National Academy of Sciences of the United States of America 101 no. 16 (2004):6315–6320. ↵

