10. Microbial Ecology and Applied Microbiology
10.1 Microbial Ecology and Metagenomics
Learning Objectives
- Define primary producer, consumer & saprophyte
- Explain the relevance of the microbes to soil and the marine environments
- Describe a Winogradsky column and its relevance to microbial ecology
- Understand the challenges of characterizing microbial communities and the various culture-independent methods that are part of the microbial ecology toolkit
- Explain why microbial ecology studies, in particular of the marine environment, are so important
- Define microbial dark matter
Microbes are vital to every ecosystem on Earth. Each species in an ecosystem is thought to occupy a separate, unique niche. The ecological niche of a microorganism describes how it responds to the distribution of resources and competing species, as well as the ways in which it alters those same factors in turn. In essence, the niche is a complex description of the ways in which a microbial species uses its environment.
The precise ecological niche of a microbe is primarily determined by the specific metabolic properties of that organism. For example, microbial organisms that can obtain energy from the oxidation of inorganic compounds (such as FeS, H2S, NH3 ) will likely occupy a different niche from the phototrophs (such as cyanobacteria). Within the euphotic zone or water column, the distribution of phototrophic bacterial and algal species are stratified according to the absorption spectra of their photosynthetic pigments and their tolerance of oxygen and H2S. Therefore, even microbes with similar metabolic properties may inhabit unique niches.
Organization of Ecosystems
Although ecologists tend to regard ecosystems as basic structural units, it can be difficult (if not impossible) to formally define the boundaries of a given ecosystem. As such, ecosystems are better thought of as conceptual rather than actual geographical locations. Rarely are ecosystems isolated from one another; rather, they should be considered parts of a larger functioning whole that together comprise the biosphere. In addition, given their microscopic sizes, within an ecosystem, conditions for the microorganisms can vary dramatically over the space of a few microns, as well as over time.
Despite the fact that clear boundaries between ecosystems may be difficult to identify, the myriad interactions that take place within an ecological community can often be observed and defined. These interactions may be best described by detailing trophic connections (what eats what) among biota in an ecosystem, thereby linking the ecosystem into a unified system of exchange.
All life forms in an ecosystem can be broadly grouped into one of two trophic levels:
- The autotrophs fix CO2, converting it into biomass. These are the producers and form the base of food chains. In terrestrial environments, the producers are the plants. In the marine environment however, the microbes are almost entirely responsible for primary production, and are either the photosynthetic or lithotrophic microbes.
- The heterotrophs are the consumers, relying on the consumption of organics that directly, or indirectly, resulted from the autotrophs. The consumers can be further subdivided into those that obtain their organics by consuming other organisms (i.e. predation), through symbiosis (pathogenesis, mutualism), or through decomposition (also called saprophytes). The latter group breaks down dead plant matter, animal material and waste products, releasing them into the ecosystem as energy and nutrients for recycling.
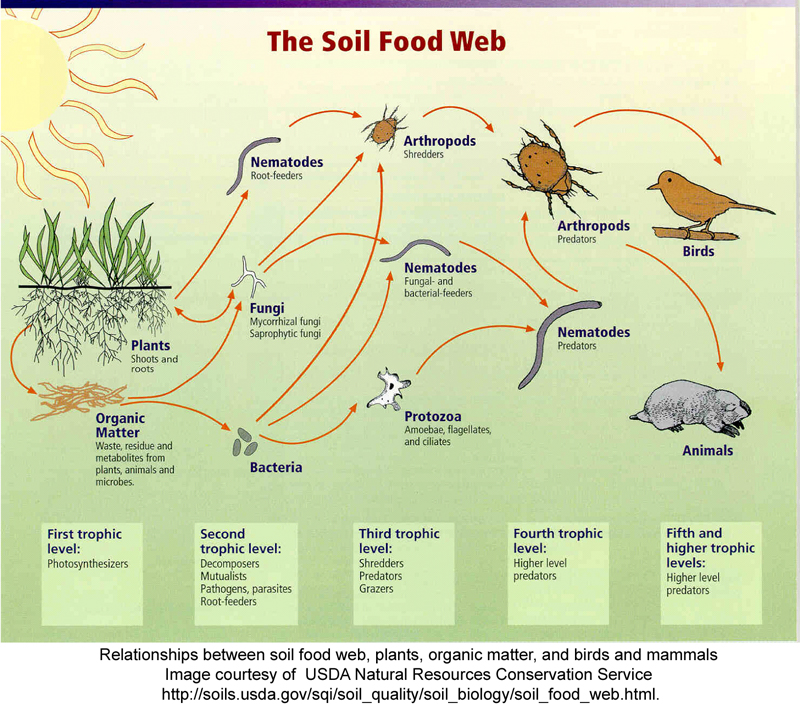
In the euphotic zone of aquatic and marine environments (i.e. the zone where light penetrates), the primary producers are largely the photosynthetic bacteria and algae. Collectively, these are referred to as the phytoplankton. Heterotrophic bacteria, archaea, protozoa and viruses all play roles in consuming or degrading cells and organics (Figure 10.3).
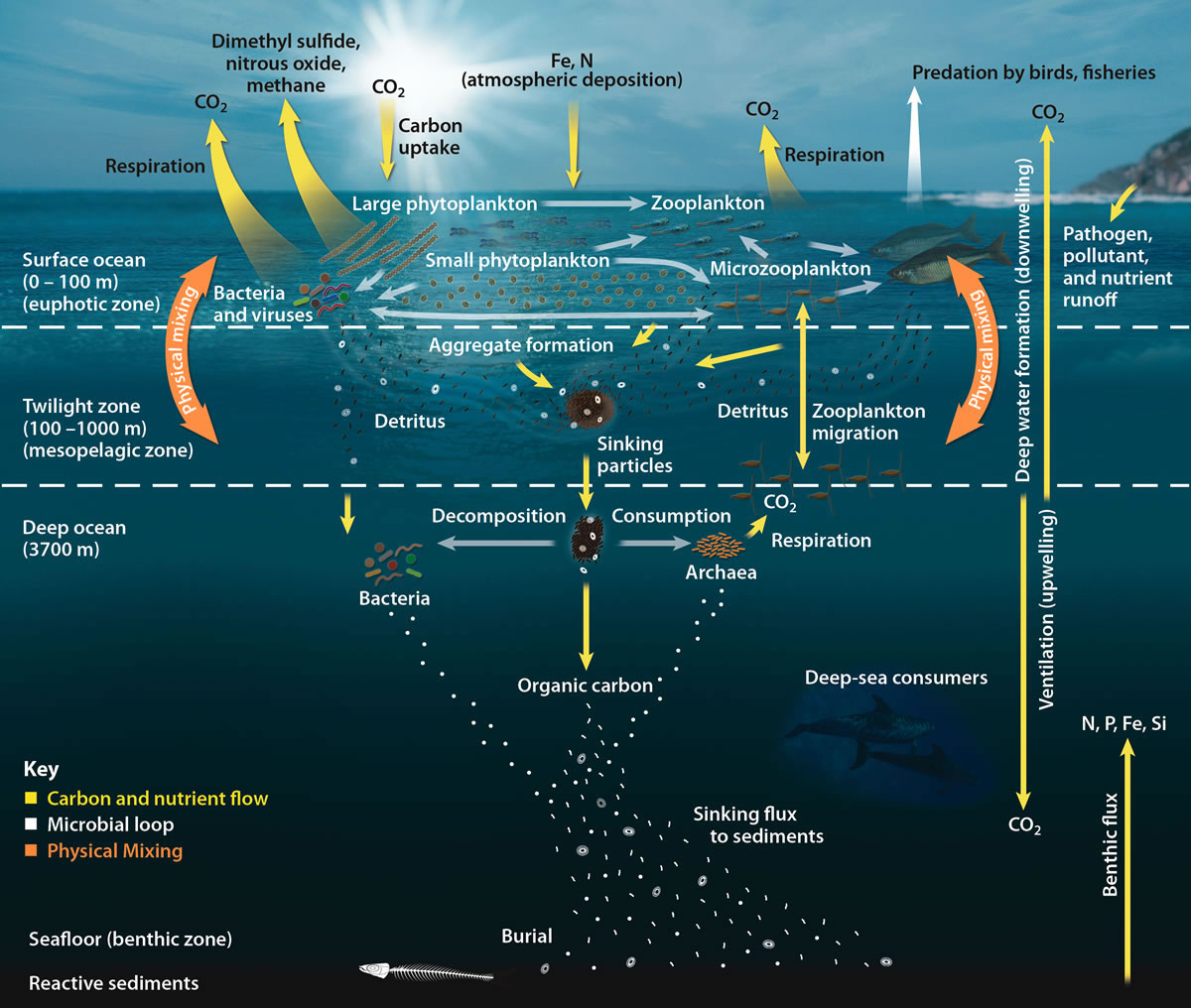
Throughout the ocean depths, where no sunlight penetrates, the lithotrophs are the primary producers. The unique food web associated with hydrothermal vents is associated with the lithotrophic bacteria and archaea. In particular, many of the lithotrophs around hydrothermal vents use the hydrogen sulphide (H2S) gas which is released from the vents, as their energy source. Some of these sulphide oxidizing lithotrophs are symbionts of the animals that grow around the vents, most notably, tube worms (Figure 10.4).

As can be seen from Figures 10.2 & 10.3, in real world ecosystems, there are multiple food chains for most organisms (since most organisms consume more than one kind of food or are eaten by more than one type of predator). Additionally, the movement of mineral nutrients in the food chain is cyclic rather than linear. As a consequence, the intricate network of intersecting and overlapping food chains for an ecosystem is more commonly represented as a food web. A food web depicts a collection of heterotrophic consumers that network and cycle the flow of energy and nutrients from a productive base of autotrophs.
Winogradsky Columns: Lab-based Microcosms
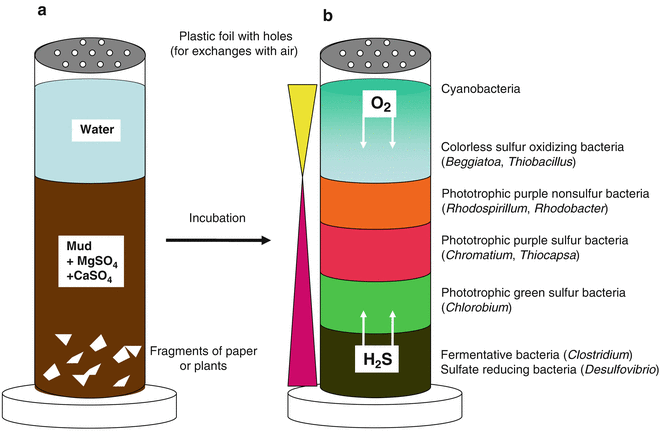
Russian scientist Sergei Winogradsky (1856-1953) was one of the first microbial ecologists. Rather than focusing on human pathogens and the isolation of pure cultures, he developed a lab-based model for wetland ecosystems: the Winogradsky column. He was the first to demonstrate that some bacteria used inorganic chemicals as an energy source. In particular, he characterized, for the first time, bacterial species that could oxidize ammonia (NH3) to nitrite (NO2–; Nitrosomonas), nitrite to nitrate (NO3–; Nitrobacter), or hydrogen sulphide (H2S) to sulphate (SO42-; Beggiatoa). Once he obtained axenic cultures of ammonia oxidizing bacteria, and H2S oxidizing bacteria, he discovered these organisms were utilizing CO2 as their sole carbon source. This previously unimagined metabolic class of organisms came to be known as the chemolithoautotrophs (or more simply: lithotrophs). Through his lab-based model ecosystems, or “microcosms”, Winogradsky also demonstrated for the first time, the critical roles of the microbes in the cycling of the elements (biogeochemical cycling). The sulphur-enriched column, or so-called sulphuretum, models the water column, with sunlight providing energy for the phototrophs, atmospheric gases diffusing in from the top, anaerobic conditions at the bottom, and partitioning of the microbes in the original inoculum according to their growth requirements and tolerance of H2S concentrations (Figure 10.5).[1] These columns are still used today, in many undergraduate (and even high school) labs.
- Who are the producers in soil and marine ecosystems?
- Who are the consumers in soil and marine ecosystems?
- What is a Winogradsky column?
Characterizing Microbial Communities
Whether trying to isolate and characterize a pathogen, or characterize the microorganisms within an environmental sample or ecosystem, the traditional strategy is to obtain an axenic culture in order to characterize the organism’s physiology and to sequence its genome. With the evolution of next generation sequencing technology, and bioinformatics, it has become possible to perform metagenomics: sequencing DNA isolated directly from the environment. Early metagenomic studies revealed that the majority of microorganisms in any environment were ones that had not previously been identified, let alone cultivated. These early studies focused on the small subunit (SSU) ribosomal RNA gene sequences: an approach based on the pioneering work of Carl Woese (see A Systematic Approach). Woese chose the SSU rRNA molecule, and its gene, because it is relatively short, present in all life forms, is not subject to horizontal gene transfer and, because it is critical to the complex and intricate process of translation, the gene experiences a relatively slow mutation rate. SSU rRNA genes therefore function as molecular clocks. These sequences are often conserved within a genus and generally different between genera. Unfortunately, the same does not necessarily apply at the level of different species within a genus. To date, there is no widely accepted definition of what exactly constitutes a “species” among the bacteria and archaea. This is particularly the case when the data is based entirely on metagenomic sequences. Regardless of this difficulty, early metagenomic surveys of SSU rRNA genes suggested that we had cultivated less than 1% of the microbial world. This is still the case today – for any environmental sample, only a small proportion of the microbes can be isolated in the laboratory.
Culture-independent Techniques
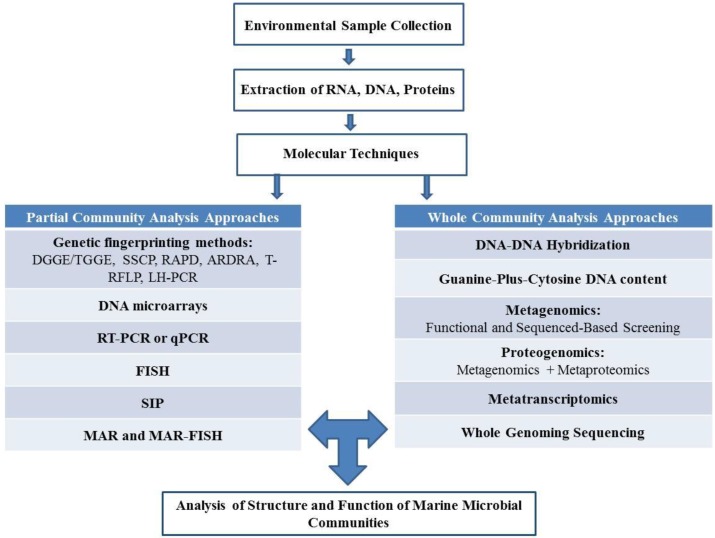
Because of the difficulty in obtaining axenic cultures, and the recognition that purified cultures behave differently then they do in environmental conditions, the field of microbial ecology now relies to a large extent on cultivation-independent techniques for characterizing community structures and ecosystem functions (Figure 10.6). In addition to using molecular techniques such as genetic fingerprinting, microarrays and PCR approaches, microbial ecologists use an increasing array of “omics” technologies that include not just DNA sequencing, but characterization of the community (RNA) transcriptome, community proteome, and even community metabolome. Beyond the molecular and “omics” techniques, the question of “who is doing what?” may be answered through a combination of imaging techniques such as fluorescence in situ hybridization (FISH; previously discussed in Staining Microscopic Specimens), microautoradiography (MAR), and stable isotope probing (SIP) to trace the incorporation of radiolabelled, and stable-isotope-labeled nutrients, respectively (e.g. 14C- or 13C-labeled organic pollutants).
Microbial Ecology of the Marine Environment and Biodiscovery
Marine metagenomic surveys have revealed unprecedented levels of diversity and led to the identification of thousands of previously unknown protein families. Given that the oceans cover approximately 70% of Earth’s surface, with depths reaching almost 11 km, this represents a veritable treasure trove of novel enzymes and products which have the potential to solve practical challenges in medicine, engineering, agriculture, and sustainability.
At the opposite end of the “omics” spectrum is the relatively newer approach of single cell genomics, obviating, to a certain extent, the need for axenic cultures, and eliminating the bioinformatic challenge of connecting metabolic function with a particular species (Figure 10.7).
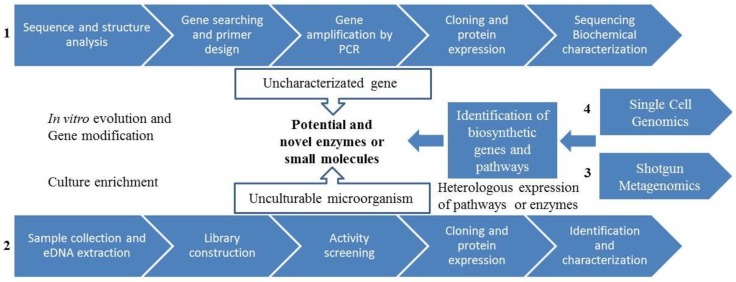
Beyond the possibilities for biotechnology and medicine, rapid technological advances are shedding light on previously unknown and uncultivated major phylogenetic groups (lines of descent) in the bacterial and archaean domains: these are the so-called “microbial dark matter”. Scientists are now able to assemble entire genome sequences from complex metagenomic DNA samples. The powerful combination of culture-independent molecular and analytical techniques has led to unprecedented advances in our understanding of microbial diversity, microbial community structure and their environmental controls. Recent investigations have even shed light on the likely role of the archaea as the host for the endosymbiosis event that led to the evolution of the eukaryotes[2]. Meanwhile, sometimes herculean efforts are leading to axenic cultures of the previously uncultivated. With axenic cultures, we can use traditional microbiological approaches for thorough characterization of novel species, thus illuminating what was previously a member of the microbial dark matter.
- What types of questions are commonly addressed when characterizing microbial communities?
- What technological advances are currently helping to answer these questions?
- What is microbial dark matter and how are we starting to illuminate these organisms?
Key Takeaways
- The microbes are important consumers in soil ecosystems, and are producers as well as consumers in marine ecosystems
- Primary producers fix CO2, converting it to biomass. Microbial primary producers are the photosynthetic bacteria and algae, as well as lithotrophic bacteria and archaea.
- Consumers are the heterotrophs, relying on organics as their carbon and energy sources. Microbial consumers may be further subdivided into predators (e.g. the protozoa), symbionts or saprophytes (the decomposers).
- Viruses are major components of marine food webs and play a major role on the marine carbon cycle.
- The Winogradsky column is a lab-based microcosm (ecosystem). It is a wetland ecosystem model developed by one of the original microbial ecologists, Sergei Winogradsky.
- The vast majority of the world’s microbes have resisted cultivation. The development of next generation sequencing techniques, combined with advances in computing and bioinformatics, have led to the use of culture-independent metagenomics as well as various other “omics” approaches in microbial ecology studies.
- Metagenomics is the sequencing of DNA purified directly from environmental samples – eliminating the need for pure cultures. SSU rRNA profiling of metagenomic DNA provides information on the types of cellular microbes present in an environment. Shot-gun metagenomic sequencing can provide information on community metabolic potential, as well as community structure (“who’s there and how many?”).
- Microbial community structure and function is now primarily examined using myriad culture-independent techniques. These include single cell genomics.
- Microbial dark matter refers to major bacterial and archaean phyla that are only now being revealed, and for which there are no cultivated representatives.
- Because the oceans represent the majority of the Earth’s ecosystems, marine microbial ecology studies in particular represent a huge potential for biotechnological and medical innovations.
- Microbial ecology studies, in particular of the marine environment, are also altering our understanding of biology, ecosystem function, and even the evolution of the eukaryotes.
Multiple Choice
True/False
Short Answer
- What is the value of sequencing the SSU rRNA from metagenomic DNA?
- What is the value of performing single cell genomics?
- What is microbial dark matter and why is it important?
Critical Thinking
- Briefly describe a Winogradsky column and explain why different microbes grow in different regions of the columns.
Media Attributions
- Soil_food_webUSDA
- Oceanic_Food_Web
- Tube worms
- Winogradsky columns
- Culture-independent commmunity analysis
- Microbial ecology discovery process
- Bertrand JC. et al. (2015) "Biogeochemical Cycles." In: Bertrand JC., Caumette P., Lebaron P., Matheron R., Normand P., Sime-Ngando T. (eds) Environmental Microbiology: Fundamentals and Applications. Springer, Dordrecht ↵
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, et al. (2017) "Asgard archaea illuminate the origin of eukaryotic cellular complexity." Nature 541: 353–358. pmid:28077874 ↵

