Team Think Tank 3 – Restriction enzyme digests
T3-3: Restriction enzyme digests.
The objective of this think tank is to help you understand the setup of a standard restriction enzyme digestion protocol. We are using resources from the company New England Biolabs (NEB) for this question, but this question can be adapted to include information from other scientific companies.
As a team, please complete the following questions. Please tackle the questions in the order they are shown.
Question 1: Using the following New England Biolabs (NEB) video as a guide: Standard Protocol for Restriction Enzyme Digests, please write out a step-by-step protocol for setting up a standard Restriction Enzyme (RE) digest reaction using the following restriction enzyme: EcoRI. The general protocol guidelines can be found here. The plasmid DNA concentration is 0.3 μg/μL and you would like to use a total of 1 µg of DNA in your reaction. You would also like to add 10 units (U) of EcoRI per reaction and you have a stock tube of EcoRI at 10, 000 U/mL.
- NEB provides different versions of the same RE. You can purchase the native/conventional RE- For EcoRI you can find the information here – or you can purchase the High Fidelity (HF) version of EcoRI (information found here). Please describe the main difference between these 2 versions. Create a table showcasing a side-by-side comparison of the EcoRI/EcoRI HF REs and the differences with respect to these REs. Here you can look at incubation time, star activity, etc. To help with this here is a great video describing star activity.
Given the provided folA and pET26b sequence information, please answer the following questions:
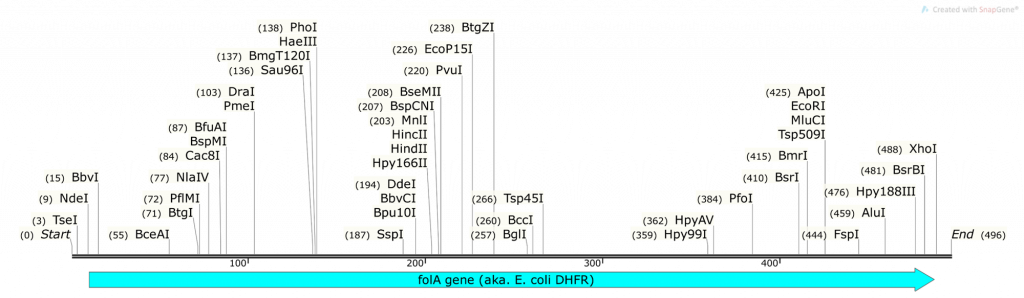
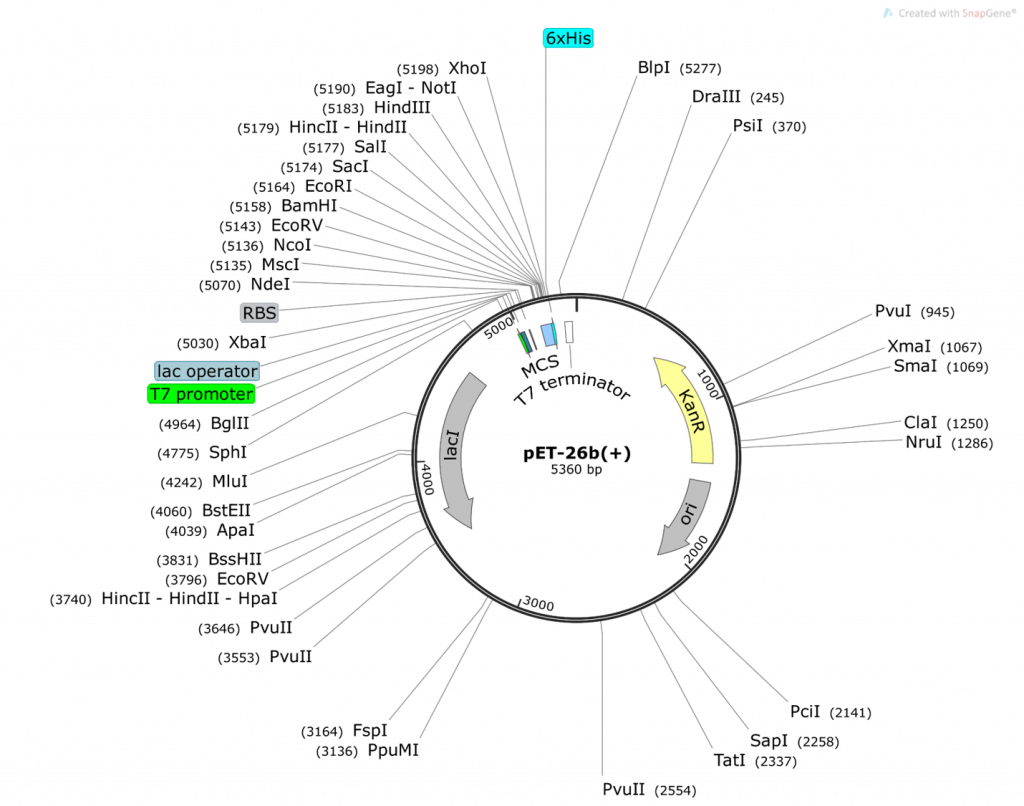
Question 2:
- What do you obtain from cutting the pET26b plasmid with NdeI and XhoI (show your math and describe ALL fragments you will obtain: tell us where the numbers are derived from)? Use the base pair positions given in the vector map as your actual cut site. Assume 100% digestion efficiency:
Question 3:
- What do you obtain from cutting the folA gene fragment with NdeI and XhoI (show your math and describe ALL fragments you will obtain: tell us where the numbers are derived from)? Assume 100% digestion efficiency. Highlight the fragment that contains your gene. Knowing that you are using the 1kb DNA ladder (see below) which of the 3 fragments obtained will you be able to clearly visualize on a 1% agarose gel?
Question 4:
- What do you obtain from ligating the two fragments together to obtain pET26b-folA? Draw the resulting plasmid (please adjust the NdeI, XhoI sizes accordingly). The plasmid must contain the T7 promoter and location of the His-tag that will be utilized in your construct (base pair positions are not necessary for these 2, just their location relative to the folA gene). The plasmid should also include the size of the folA gene. What is the size of your newly constructed plasmid? Please show your math and clearly explain how you obtained your final plasmid.
- On your pET26b-folA plasmid diagram, please highlight the new bp positions for PvuI (both in the plasmid and in the gene) and explain how you achieved these numbers (please show your math!)
Question 5: Please create a hypothetical agarose gel result containing the outcomes of your double restriction digest for the following samples:
-
-
- 1kb DNA ladder
- pET26b plasmid cut with NdeI/XhoI
- pET26b plasmid cut with PvuI
- pET26b-folA plasmid cut with NdeI/XhoI
- pET26b-folA plasmid cut with PvuI
- pET26b-folA plasmid cut with PvuI/ NdeI/XhoI.
- pET26b-folA plasmid cut with PvuI/ PvuII.
-
At the bottom of the gel you need to state each fragment (for each lane) and describe how you obtained this number (show your math). If you have already calculated band sizes in previous questions, please refer to that question. You do not need to recalculate.
The hypothetical agarose gel image is provided. Please use this gel template to add your bands.


