Chapter 5 Lab Overview and Background Information
Learning Outcomes: Upon completion of this lab, students will gain practical experience in:
- Selecting restriction enzymes for diagnostic purposes
- Loading DNA samples onto an agarose gel
- Interpreting DNA agarose gels
In this lab you will be using the same techniques introduced in chapter 2, but for a slightly different purpose. Let’s recap:
Our goal thus far has been to clone the E. coli folA gene downstream of the T7/lac promoter in the pET26b plasmid. Following the ligation reaction and subsequent bacterial transformation and alkaline lysis miniprep, we now have a micro-centrifuge tube with purified plasmid DNA … but did we achieve our goal? As discussed in chapter 3, the bacterial colonies should contain our plasmid, but we have no way of knowing if our folA gene was successfully cloned in the plasmid. In fact, we are probably looking at two possible scenarios in our purified plasmid tube. Scenario 1, which is our desired result, or scenario 2, which is our undesired result.
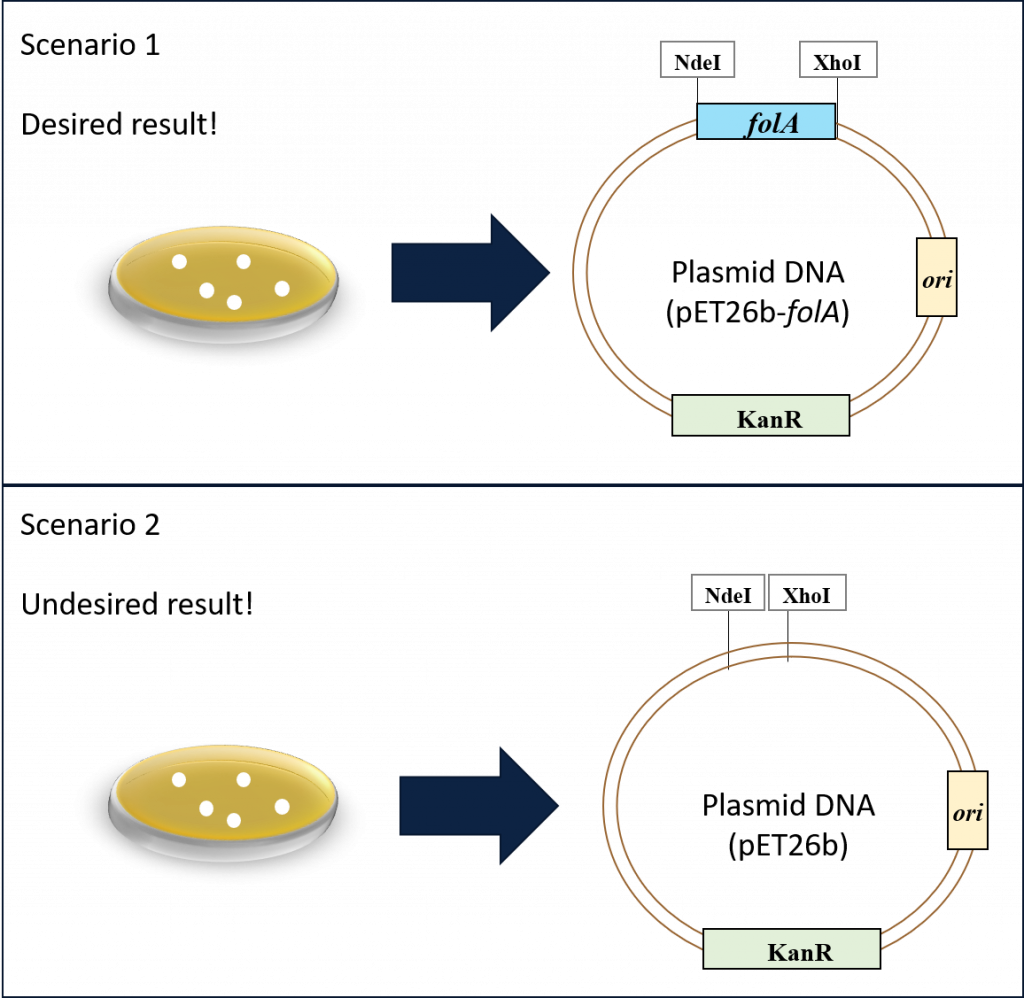
Ans so, it’s time to get creative and repurpose already learned techniques for new uses. We will start with REs. The first time we used REs to undergo directional cloning of our folA gene into the pET26b plasmid. This time around we will use REs to solve a DNA puzzle. We already know that REs “cut” at specific DNA sequences (aka. palindromes), and we can use molecular cloning software to predict exact DNA fragments following RE digestion reactions. And so, we can use these features to create unique DNA fragment “fingerprints” specific to each of our two DNA plasmids in the above scenarios.
The workflow is as follows:
Restriction enzyme mapping as a means to verify your purified plasmid
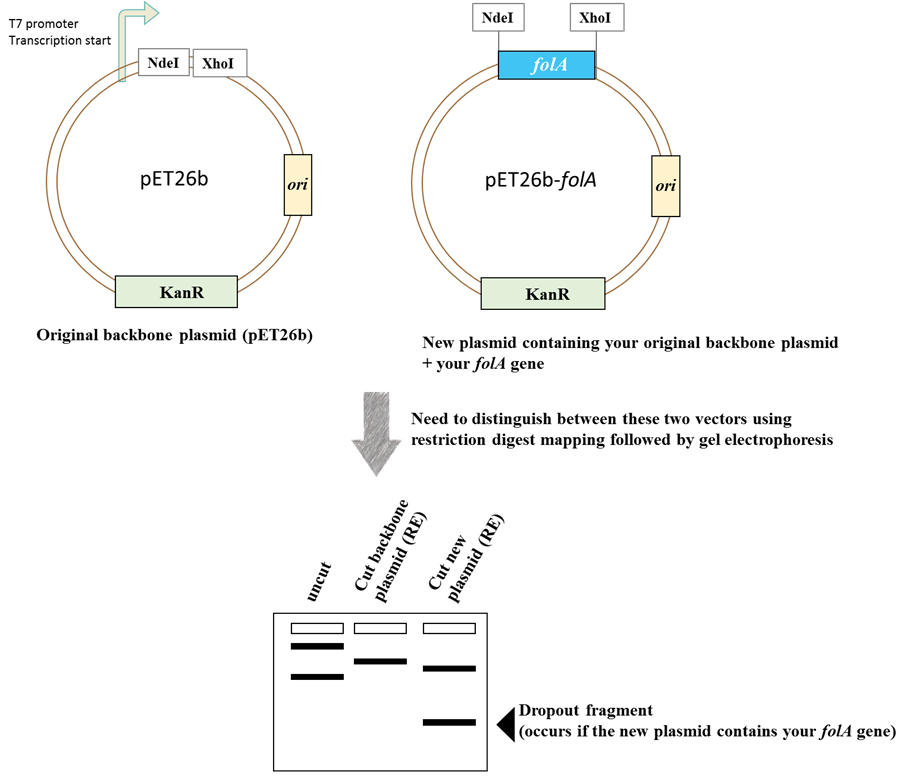
Image created by Felicia.
Schematic diagram of the restriction enzyme digests mapping strategy for determining the presence of folA in pET26b.
Want to learn more about RE mapping? Check out the module here

