50 4.3: ChIP Sequencing
Chromatin immunoprecipitation (ChIP) is a procedure used to analyze protein interactions with DNA. Using ChIP sequencing, one can map out binding proteins and histone modifications down to each base pair. It is considered a next-generation piece of technology for genome-wide sequencing. It is able to generate large amounts of data at a time and allows for detailed analysis of DNA.
How It Works
ChIP uses antibodies to select for specific proteins and nucleosomes by immunoprecipitating the DNA-protein complex. This process enriches the DNA being studied. Oligonucleotide probes are placed across the genome or specific promoter regions of it at a high resolution. It is then observed through microarray technology. ChIP-seq is a newer version of its previous counterpart ChIP-chip microarray. In ChIP-seq, DNA fragments are sequenced directly instead of being hybridized on a microarray like ChIP-chip. (Park)
ChIP-seq and LIF
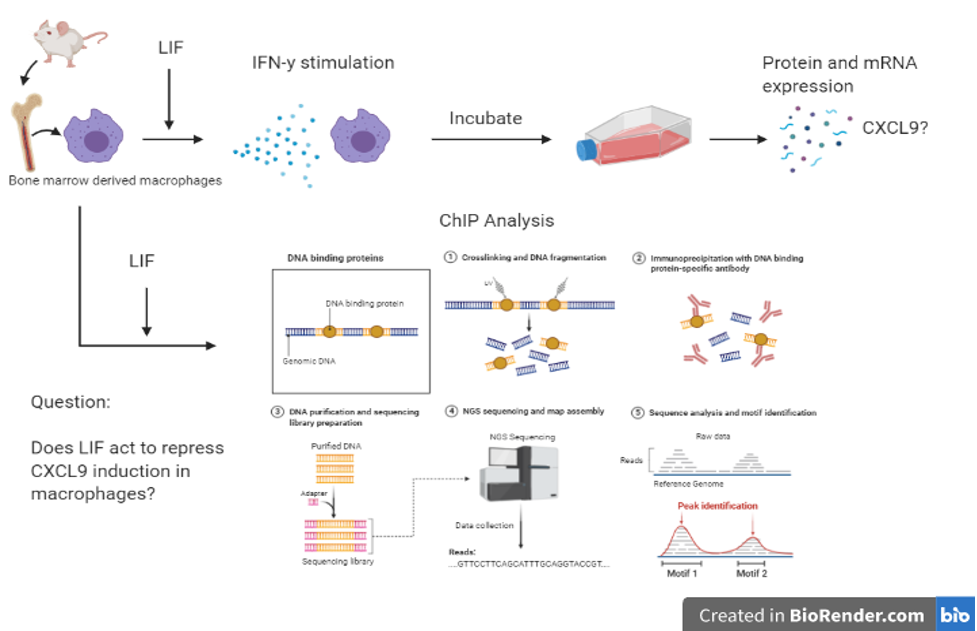
The paper by Pascual-García, et al. (2019) mentioned in the previous chapter also used ChIP sequencing in their study. The researchers wanted to look at the relation between LIF and the cytokine CXCL9. They first treated bone marrow derived macrophages with LIF. They then did a ChIP analysis of Tri-methylhistone H3 (H3K27me3), EZH2 and acetyl-histone4 (H4ac). The results indicated that treatment with LIF increased levels of Tri-methylhistone H3, decreased acetylated-histone 4, and increased binding of EZH2 to the CXCL9 promoted. This meant that LIF played a role in the epigenetic silencing of CXCL9 and ultimately the inhibition of CD8+ T cell tumour infiltration.
Test your knowledge:
