17 Introduction to Metabolism: Metabolic Pathways
The chart in Figure 17.1 is a schematic of cellular metabolic pathways. We are going to study only a very small amount of this complexity. Our goal; however, is not just to memorize facts (although we need to do some of that), but to draw, from just a few specific examples, general principles that we can apply to all aspects of biochemistry. As we learn about metabolism, we soon see that there are many common themes to cellular chemical transformations, whether we are considering processes as diverse as amino acid biosynthesis, steroid hormone metabolism, or DNA repair.
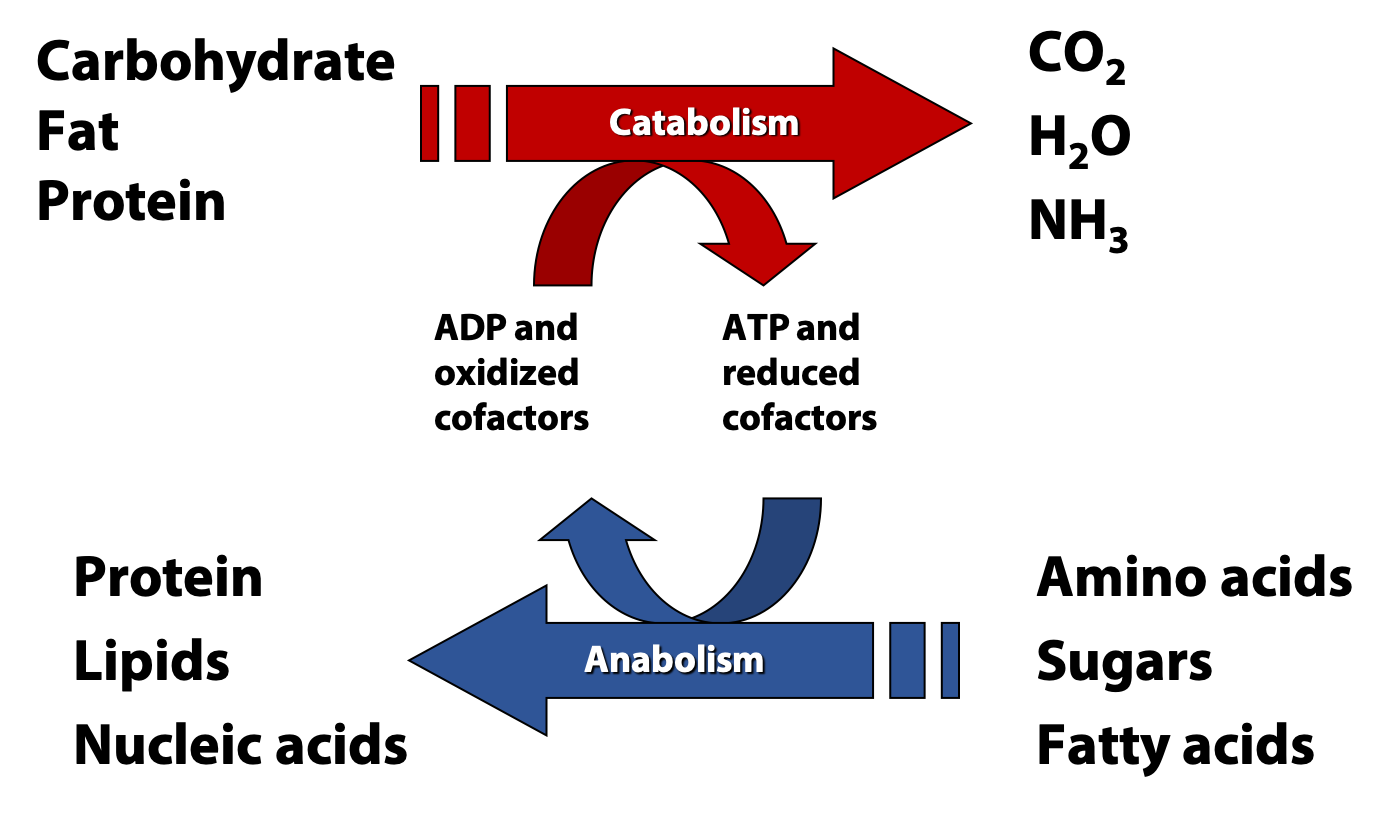
We can divide metabolism pretty cleanly into two halves: anabolism, the process of building up larger and more complex molecules from simple precursors, and catabolism, the breakdown of large molecules and foodstuffs into simpler products (Figure 17.2). In this course, we are going to focus on the central catabolic pathways of fatty acid and sugar metabolism.
All cells are carrying out both catabolic and anabolic processes simultaneously. Between these two halves of metabolism are a myriad of connections involving compounds, reactions, genes, cellular physiology, and energy. In most cases, catabolic processes supply ATP energy and anabolic processes require that energy. Energetically “uphill” reactions in anabolism are usually driven forward by coupling to the hydrolysis of ATP, as we have already discussed.
Catabolic pathways converge
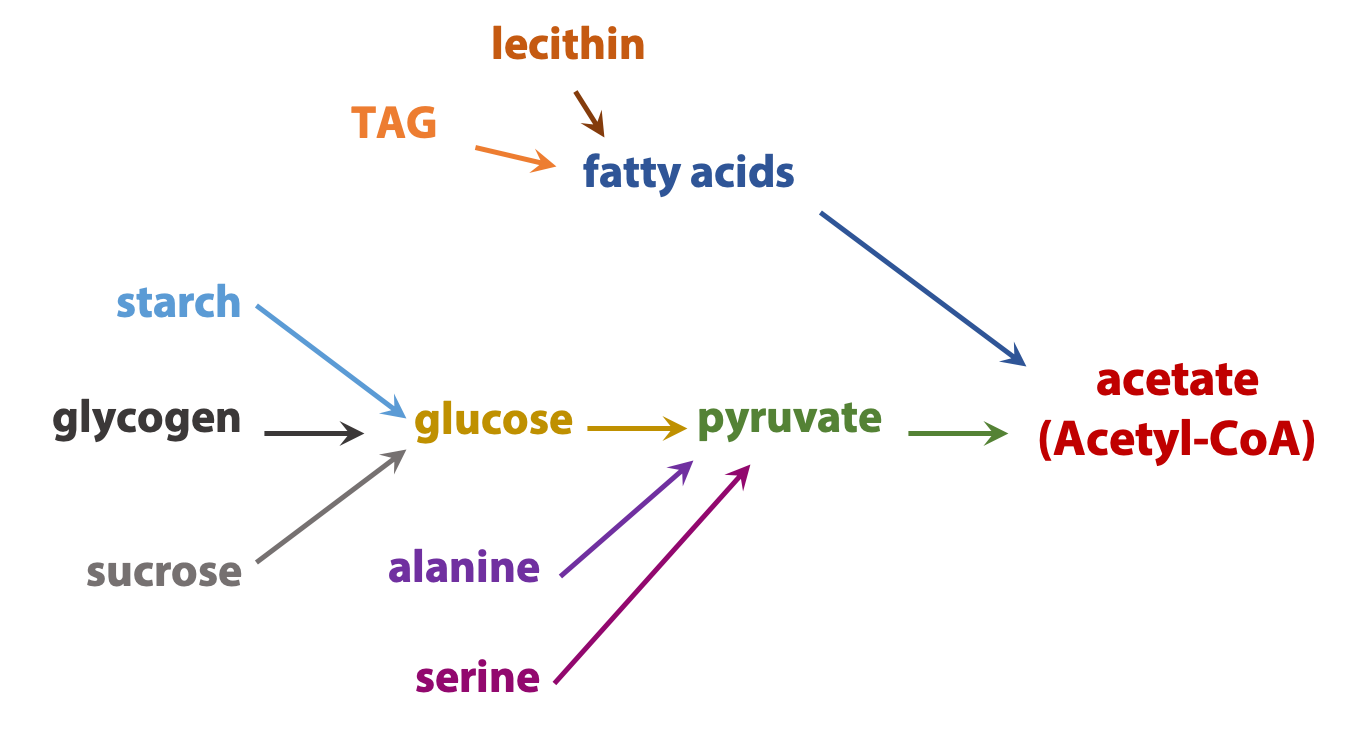
The complexity of metabolism is undeniable; even the simplest cell contains hundreds of small molecules and thousands of proteins. But many common themes simplify this complexity. Here is one theme: Catabolic processes usually converge. For example, the carbon “skeletons” of most sugars, fats, and amino acids are converted into a single centrally-important metabolite, a carrier of two-carbon (acetate) units called acetyl coenzyme A (Figure 17.3). This pattern allows the cell to operate by using a rather limited number of high-flux carefully regulated “core business operations”, while retaining the metabolic flexibility to make use of a wide variety of chemical “feedstocks”.
Anabolic pathways diverge
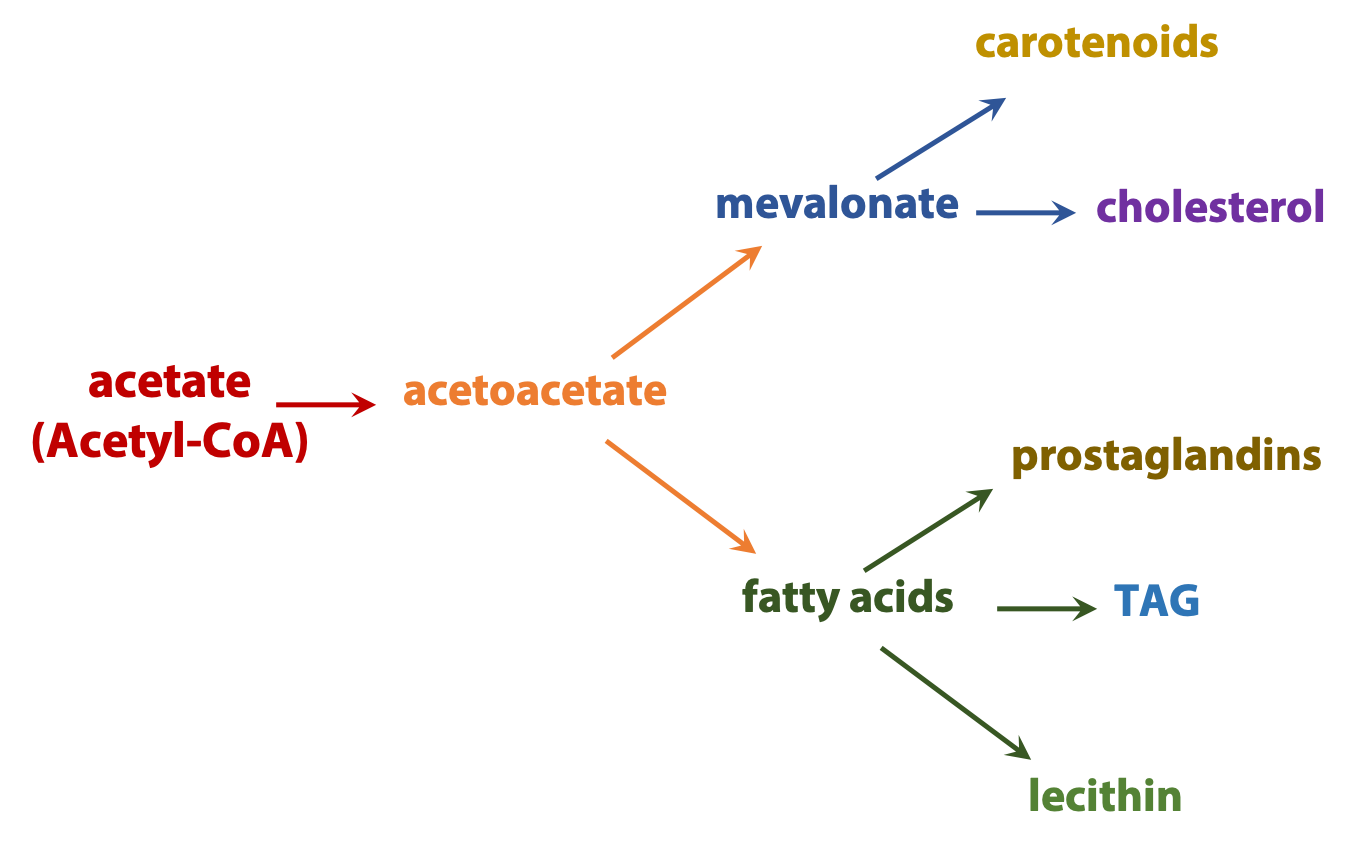
Anabolic processes diverge. For example, acetyl coenzyme A can be used to build everything from fatty acids to steroids or components of proteins and nucleic acids (Figure 17.4).
Characteristics of metabolic pathways
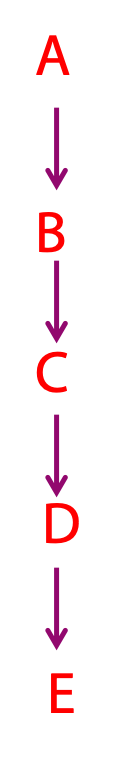 Even the most complex metabolic pathway, for example, converting a starting material into something unrecognizable such as going from glutamate to heme occurs one step at a time. Each step is an enzyme-catalyzed chemical reaction. Many of these reactions follow simple and easily-understood chemical avenues, and the same chemical “themes” occur over and over again, in slightly different contexts. Once you can recognize a few of these chemical “motifs”, the complexities of metabolic pathways begin to seem less bewildering. These reactions convert a precursor (A, at right) into a product (E) through a series of intermediates known as metabolites. Each step in a metabolic pathway brings about a small but specific chemical change (the removal, addition or transfer of a particular atom or functional group). While each step is strung together in order, it can bring about a transformation that is not at all obvious.
Even the most complex metabolic pathway, for example, converting a starting material into something unrecognizable such as going from glutamate to heme occurs one step at a time. Each step is an enzyme-catalyzed chemical reaction. Many of these reactions follow simple and easily-understood chemical avenues, and the same chemical “themes” occur over and over again, in slightly different contexts. Once you can recognize a few of these chemical “motifs”, the complexities of metabolic pathways begin to seem less bewildering. These reactions convert a precursor (A, at right) into a product (E) through a series of intermediates known as metabolites. Each step in a metabolic pathway brings about a small but specific chemical change (the removal, addition or transfer of a particular atom or functional group). While each step is strung together in order, it can bring about a transformation that is not at all obvious.
Metabolic pathways are “one way streets” (irreversible) i.e., they always occur in one direction. They contain at least one reaction that is thermodynamically very favourable. All metabolic pathways are regulated and the regulation takes place near the beginning of the pathway. Activity of enzymes are regulated through transcriptional control of the level of enzyme as well as through inhibition and activation of the activity of the enzyme e.g., through reversible phosphorylation, feedback inhibition.
Precursor-product relationship
The elucidation of complex metabolic pathways can be regarded as the identification of a series of linked precursor-product relationships, in which the product of one step is the precursor for the next. None of the steps is, by itself, remarkable. However, when strung together in order, they may effect a transformation that is not at all obvious.
Elucidation of metabolic pathways
Cells are very complex. Thousands of reactions are happening at the same time. A few metabolic pathways have huge fluxes, which means very large amounts of material are being processed through these pathways. These are mainly the “central” catabolic pathways, like glycolysis. In fact, they were the first pathways elucidated in the early 20th century, by scientists using techniques that seem very crude and low-sensitivity (by today’s standards). For example, most of glycolysis was figured out by simply taking cells, like yeast, making a cell-free extract, adding glucose, and then using old-fashioned chemical analysis methods to detect the products. This approach didn’t work very well until Harden and Young discovered (completely by accident, like a lot of good discoveries!) that the process goes much faster when phosphate is added. After that, they were able to figure out the products. However, if the metabolic pathway has a smaller flux, the amounts of product are so small that people can’t easily detect them by “low-tech” methods. As a consequence, a lot of pathways remained completely mysterious. Take the steroid cholesterol, for example. By the 1930s, its structure was known and everyone realized that it was a very important metabolite – but nobody knew anything about how it was made in the cell.
The elucidation of a metabolic pathway begins with the task of figuring out the precursor-product relationships: which compound is converted into which compound, step by step? Then, we can ask about the stoichiometry of each step – which other cofactors and substrates participate in the reactions? What are the enzymes that catalyze each step? How are the steps regulated? Which genes encode them? All of these questions are of interest to biochemists.
Experimental approaches to the study of metabolic pathways
- The use of metabolic inhibitors
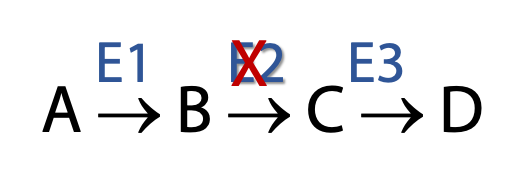
Figure 17.5 Using an inhibitor to E2 to determine the order of enzyme reactions in a pathway Cells are very complex. Thousands of reactions are happening at the same time. Only a very few metabolic pathways have large enough fluxes through them to allow us to isolate and identify the intermediates without any external manipulations (e.g., glycolysis). Flux through most metabolic pathways is very small such that isolation of specific intermediates from among the thousands of metabolites present in a cell is quite impossible.
Metabolic inhibitors block pathways at specific points (Figure 17.5). This leads to a buildup of preceding intermediates. As shown in the example above, if the enzyme E2 is inhibited, B begins to accumulate as it can no longer be converted to C. The presence of increased amounts of B enables us to isolate and identify it.
- Biochemical Genetics
a) Genetic Diseases

Figure 17.6 The deficiency in a specific enzyme in a pathway due to a genetic change leads to the accumulation of metabolic intermediates. In 1940, George Beadle and Edward Tatum appreciated that each enzyme in a cell is encoded by a specific gene. The fact that human genetic diseases are a consequence of deficiencies in specific enzymes was first identified by Archibald Garrod in 1904. These “experiments of nature” also have made a large contribution to our understanding of metabolic pathways. The effect of genetic diseases is similar to that described above for the use of metabolic inhibitors. The deficiency of a specific enzyme of a metabolic pathway leads to the accumulation of intermediates preceding the point of deficiency (Figure 17.6). e.g., individuals with alkaptonuria, a disease arises due to a deficiency of an enzyme in the pathway of phenylalanine catabolism, which causes to accumulate homogentisic acid in their blood and eventually leading to its excretion in the urine. These patients lack the enzyme that breakdown homogentisic acid.
b) Auxotrophic mutants
Escherichia coli (the bacterium we most often study in the lab, discovered by Dr. Theodor Escherich) is a prototroph (“first – food”). All it needs is sugar, a nitrogen source like ammonia, some mineral salts and it will grow happily, making every metabolite it needs. We call that recipe of sugar and salts is “minimal medium”. But if a gene encoding an essential enzyme is inactivated by a mutation and the strain can’t make the product of a particular metabolic pathway, it will become an auxotroph (“help – food”).
Auxotrophs can be produced by exposing a bacterial culture to a mutagen to induce mutations in its genes. Bacteria carrying mutations in enzymes of a specific pathway can be identified by their requirement of the end product of that particular pathway for growth. i.e., if there is a mutation in an enzyme in the pathway of proline synthesis, the mutant strain is unable to make the amino acid proline. It is no longer a prototroph and requires a supply of proline for growth. This mutant strain can’t grow on minimal medium, but it can grow on minimal medium plus proline! The bacteria can just take up the proline that they need from the medium. (The same would be true for lots of other metabolites; proline is just used here as an example). Once we find such a mutant, we can analyse it chemically/biochemically, to identify metabolites that build up to unusually high levels – they are probably precursors of proline. Then, we can analyse it genetically to find the gene that is mutated – it must be a gene encoding a proline biosynthetic enzyme.
Finding auxotrophic mutants by replica plating

How do we find such auxotrophic mutants? We can do this systematically. Firstly, we take an E. coli culture and expose it to a mutagenic chemical to induce mutations in many of the bacterium’s genes. Next, we plate out the cells onto minimal medium supplemented with proline and let each cell grow up into a colony (that takes about 48 hours). Then, we take a piece of velvet cloth, sterilize it in an autoclave, and gently press it onto the petri dish. The surface of the velvet picks up a little bit of each colony. Next, we press the velvet onto a second petri dish (Figure 17.7). This second dish has minimal medium only – it is not supplemented with proline. All the transferred cells form colonies, so the second plate is a “replica” with the identical pattern of colonies.
-
The use of radioactively labeled substrates
Large amounts of radioisotopes, such as 14C, can be made by bombarding a target with neutrons. 14C behaves just like regular carbon (12C), in terms of its chemistry, except the radioisotope can be “traced” easily, because when it decays, it releases a little flash of energy. This invention enabled biochemists to study the metabolic pathways that have very small fluxes through them (which means there are very small amounts of the intermediates).
If you synthesize a compound – e.g. methionine – from 14C, then the compound and any products derived from the carbon atoms in it, will be radioactive. As a result, they can be easily detected, with techniques like scintillation counting or autoradiography. Moreover, it will work even if the methionine in a cell is surrounded by thousands of other (non-radioactive) compounds. Suddenly, it became possible to trace biochemical products.
Self-assessment Questions
1.
2.
