1 Biological Macromolecules and Amino Acids
Synopsis: Molecules of interest to biochemists may be classified as small molecules and macromolecules. Small molecules are similar to those encountered in conventional organic chemistry. They are important in metabolism, which we deal with in the second half of the course. Macromolecules are huge by comparison – molar masses from 104 to over 109 g.mol-1. What makes it possible to comprehend structures of this magnitude is their modular construction from much simpler smaller molecular units. The basis of macromolecule assembly is the reversible formation of certain kinds of bonds, e.g. ester or amide bonds to link up smaller subunits into long chains. Proteins are chains of linked amino acids. Each amino acid has a unique side chain. Since the α-amino/α-carboxylate core is constant, the side chain R determines the specific properties of a particular amino acid and the role it plays in a protein.
REVIEW: CHEM*1040 notes regarding electronegativity and Lewis structures.
Classes of molecules found in biochemistry.
1. Small molecules
- Sugars, amino acids, nucleotides, fatty acids, simple carboxylic acid derivatives
- Interconversions of small molecules may be used to store or release energy, which is the basis of metabolism.
- Particular kinds of small molecules may serve as building blocks for macromolecules.
2. Macromolecules
- Proteins, made as chains of amino acids
- Nucleic acids, made as chains of nucleotides
- Polysaccharides, made as chains of simple sugars
Proteins form complex structures capable of many functions, including structural components of cells, catalysis of reactions and communication processes. For this reason, the first half of the semester will focus on proteins and their role.
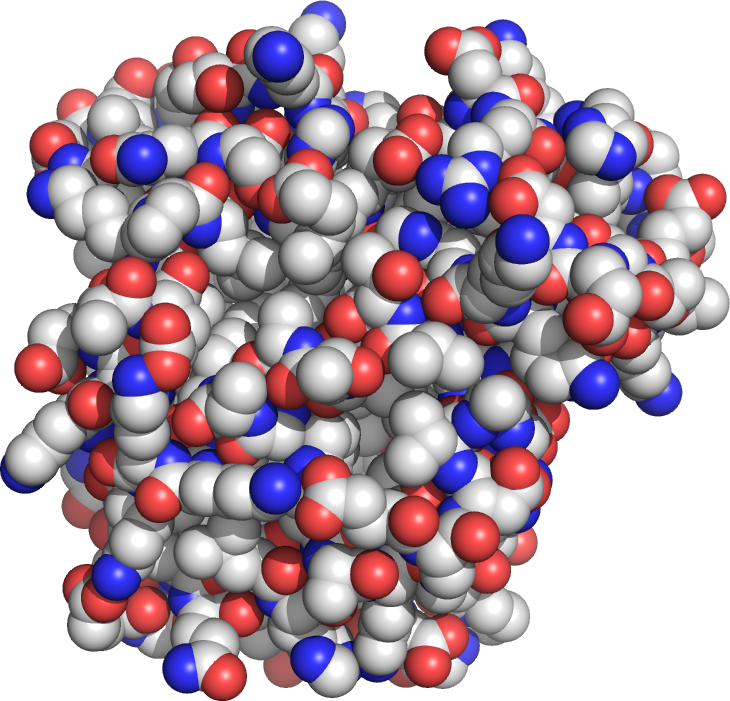
For example, myoglobin (Figure 1.1) is a protein that stores O2 in muscle tissue. Typical protein molecules have molecular masses between 10 000 and 100 000 g.mol–1, so they contain literally thousands of atoms. Myoglobin has a molecular mass of 16 500 g.mol–1.
Because proteins and other macromolecules are so large, biochemists use a unit called the kiloDalton (kDa). One Dalton is simply 1 g.mol-1, so 11 000 g.mol–1 becomes 11 kDa. Typical proteins are between 10 and 100 kDa, while myoglobin is 16.5 kDa. The largest known single protein molecule is titin at 10 000 kDa!
The building block principle of macromolecule structure.

Proteins are chains of linked amino acids (Figure 1.2): Each protein has a unique sequence of different amino acids, and a well-defined size and structure. The arrangement of amino acids in the chain determines the properties and function of the protein. A protein of 100 amino acids has a mass of about 11 000 g.mol–1, about 110 g.mol–1 per amino acid. Proteins are between 10 and 10 000 kDa (104 to 107 g.mol-1).
Two other kinds of macromolecule that will be dealt with later in the course:

1. A polysaccharide is a chain of sugars (Figure 1.3):
Most polysaccharides, e.g. starch, are simple repetitive structures of one or two sugars, with no definite size. Some polysaccharides are used for storage of sugars. Others act in simple structural roles.
2. Nucleic acids DNA and RNA:

The backbone is simple and repetitive, but different bases are attached giving nucleic acids unique and characteristic sequences (Figure 1.4). The repeating unit, base + sugar + phosphate is called a nucleotide.
Bonding between subunits.
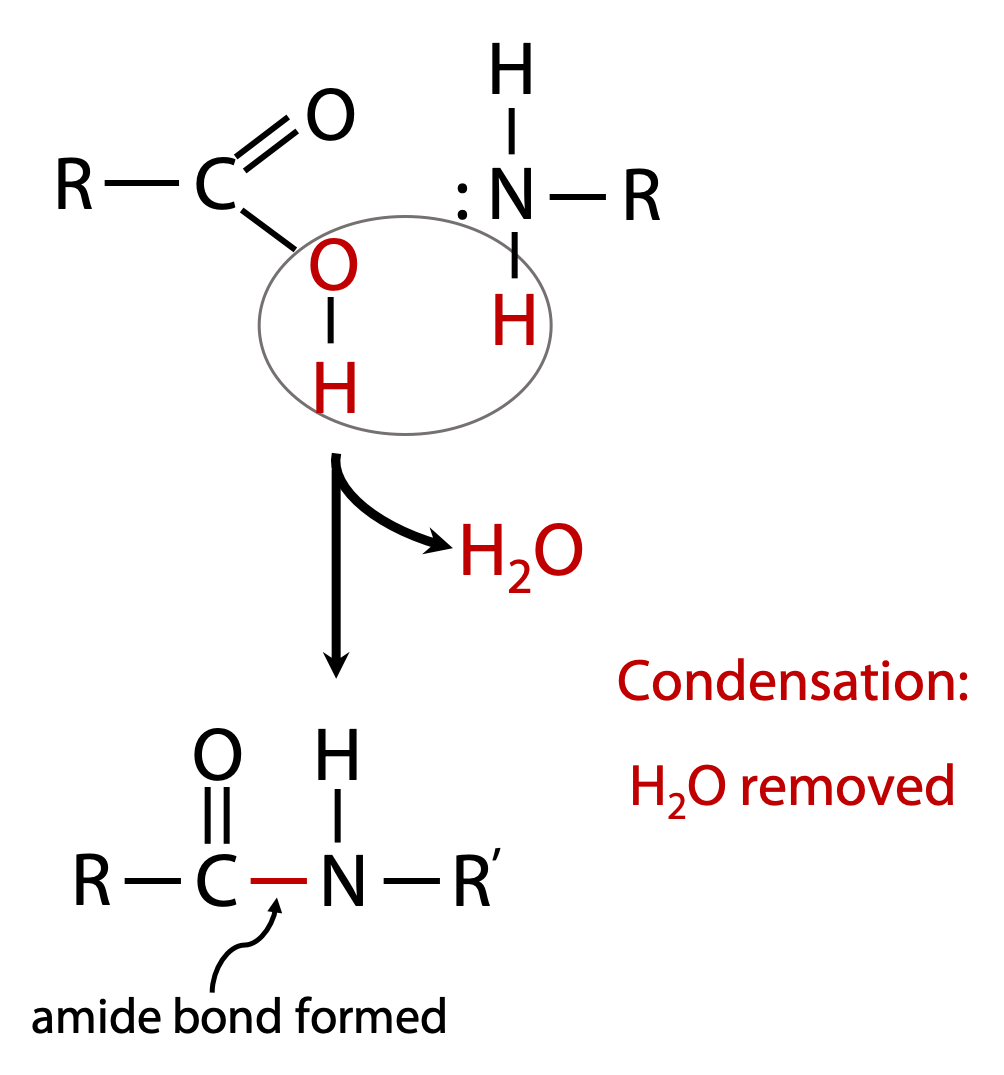
The types of bond that link subunits in macromolecules are formed by a process called condensation, since the process involves elimination of the elements of H2O (Figure 1.5).
e.g amino acids contain both carboxylic acid and amino groups, and these allow the formation of an amide bond by condensation:
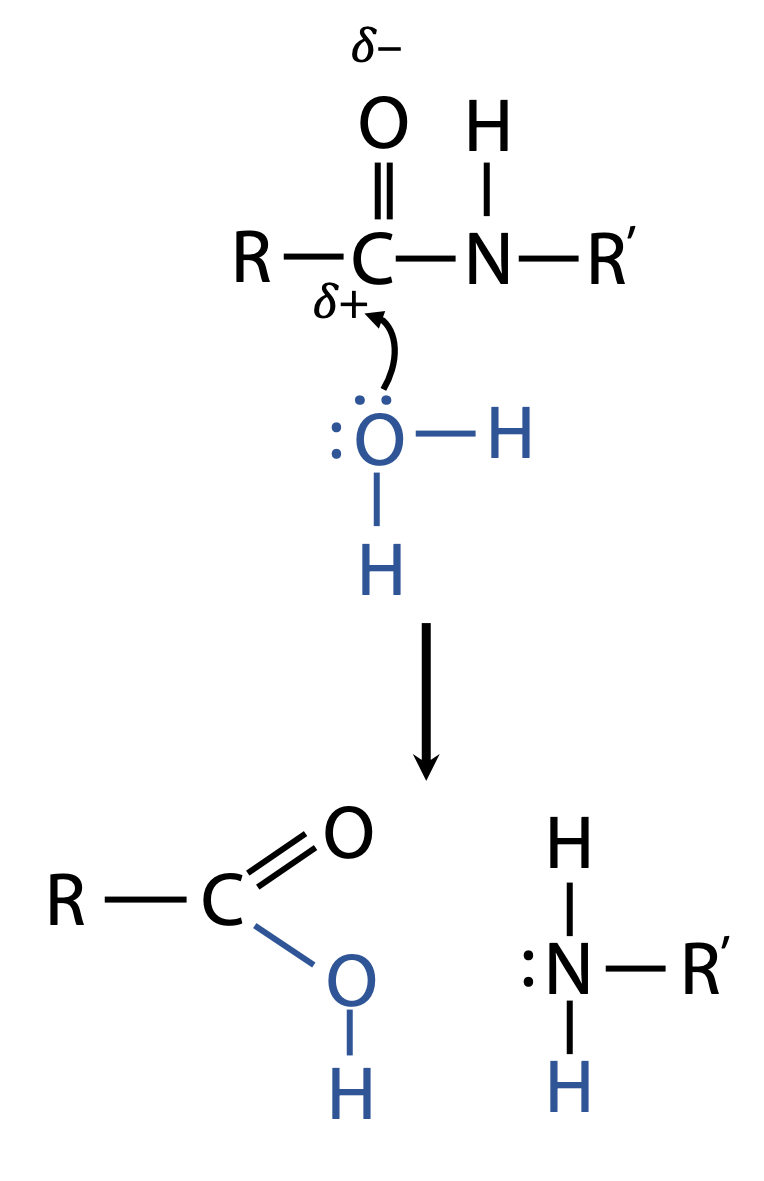
The converse of condensation is the attack of H2O on the amide bond, which restores the original carboxylic acid and amino group. Thus, it unlinks the two units. This is called hydrolysis (Figure 1.6).
The carbonyl group C=O of the amide is the point of weakness that allows H2O to attack.
Bonds formed by condensation and broken by hydrolysis:
carboxylic acid + amino group = amide
carboxylic acid + alcohol = ester (Figure 1.7)
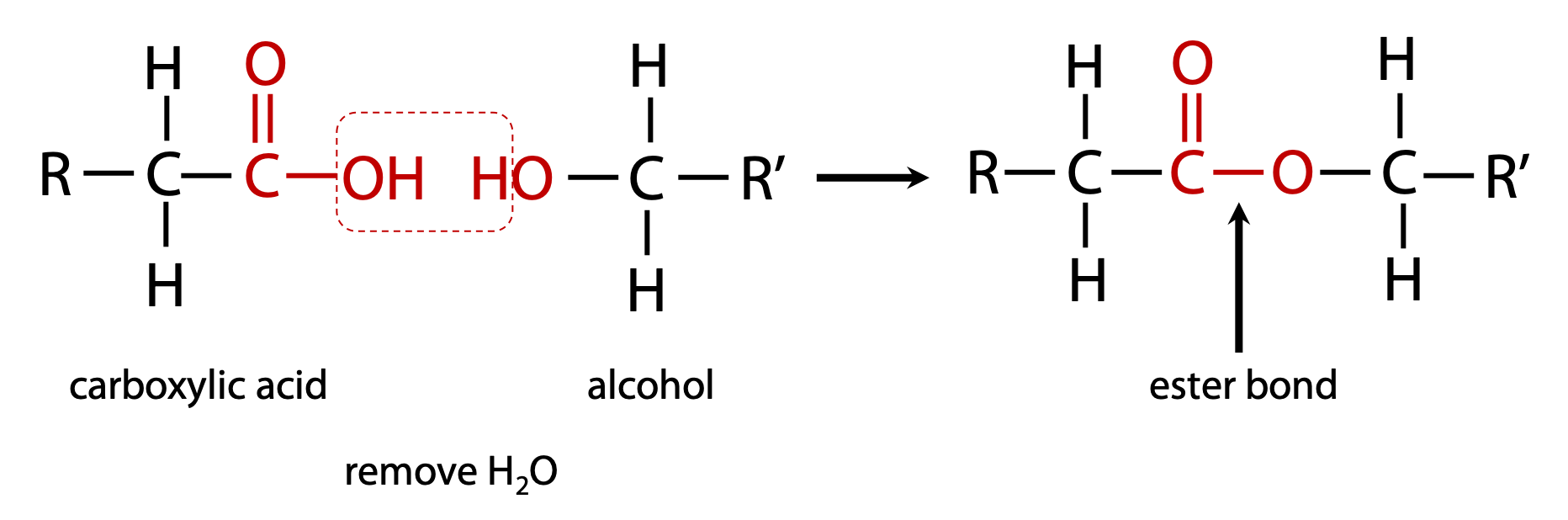
Proteins are chains of amino acids

Twenty different amino acids are found in proteins. They have in common the structure in Figure 1.8, distinguished by different side chain groups, shown as R.
Each amino acid has an amino group and a carboxylate group; therefore, it can form both end of an amide bond. (What’s different about the amino acid proline? – look it up and find out!)
After removal of the H2O in condensation, the portion of the amino acid found in the chain is called an amino acid residue (Figure 1.8).

Large numbers of amino acids can be strung together in a chain. Each amino acid in the chain can have a different side chain R. These give the protein molecule as a whole its unique properties (Figure 1.9).
On average, each amino acid contributes about 110 Da to the molar mass of a protein. 153 amino acids make up the 16.5 kDa myoglobin molecule.
Some terminology to learn:
One end of the chain has an amino acid with an uncombined amino group, usually protonated. This is called the N-terminal amino acid. The other end has an amino acid with an uncombined carboxylate group, which is called the C-terminal amino acid (Figure 1.9).
Amino acid chains linked in this way are called peptide chains. The amide bonds linking them are called peptide bonds.
A polypeptide is a large peptide chain, usually the complete amino acid chain of a protein. Typical proteins are several hundred amino acids long.
Poly = Greek for “many”.
An oligopeptide is a smaller peptide chain, often applied to a fragment of a larger protein.
Oligo = Greek for “a few”
Structural layout of amino acids
Amino acids in a peptide chain have identical backbone but unique side chains R. Specific properties of a protein are determined by the particular functional groups present in the side chains. The amino acid glutamate (Glu for short) is an example.
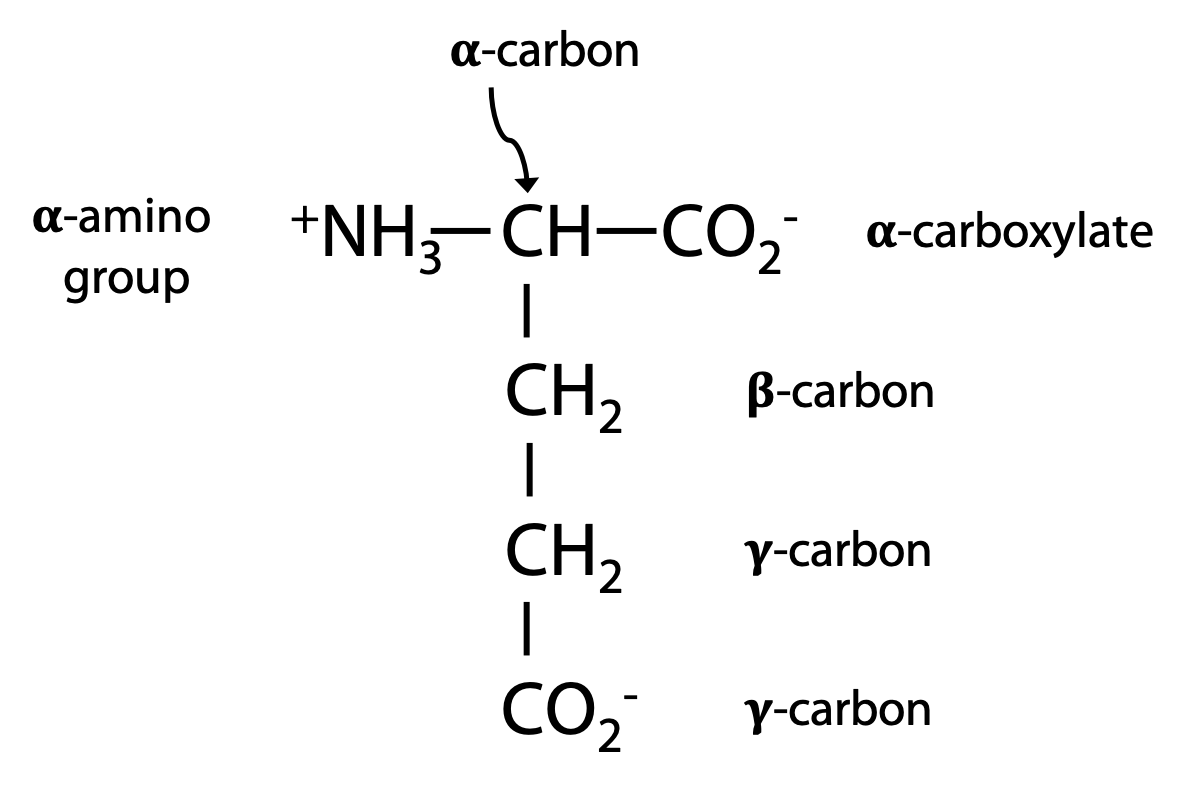
In biochemical nomenclature, Greek letters identify the carbon atoms of the structural core of an amino acid, as in the example at left:
Functional groups are labeled according to the core atom to which they are attached, e.g. α-amino, α-carboxylate, γ-carboxylate. The α-amino and α-carboxylate become linked up in the peptide bonds making up the backbone (Figure 1.10).
The side chain properties that have most influence on the behaviour of the protein include:
- polarity
- hydrogen bonding ability
- charge
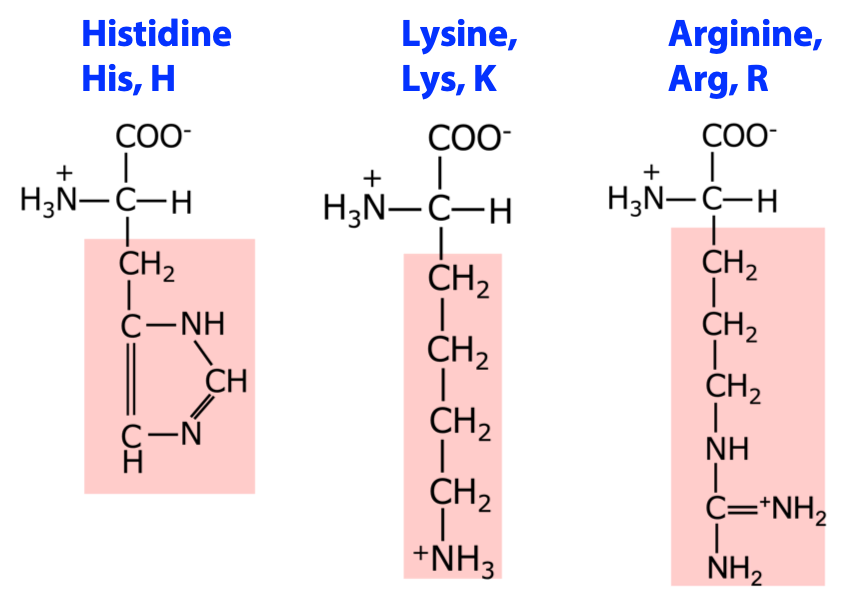
There are twenty different amino acids commonly found in protein chains, described by a full name, a three-letter abbreviation, or a single letter symbol. Full structures of amino acids and other properties can be found in the Stryer textbook (click here for the link) and on the amino acids Euchre deck (available online).
Note: You should be prepared to reproduce the structures of the amino acid constituents of proteins and the complete covalent structures of proteins; to know the single- and three-letter abbreviations for the amino acids.
Classification of amino acids by properties
The “inverted pyramid” below is a convenient memory aid which groups amino acids according to common properties and structures. Know these classifications, the structures of the amino acids, their single- and three-letter codes.
6 amino acids with very non-polar side chains:
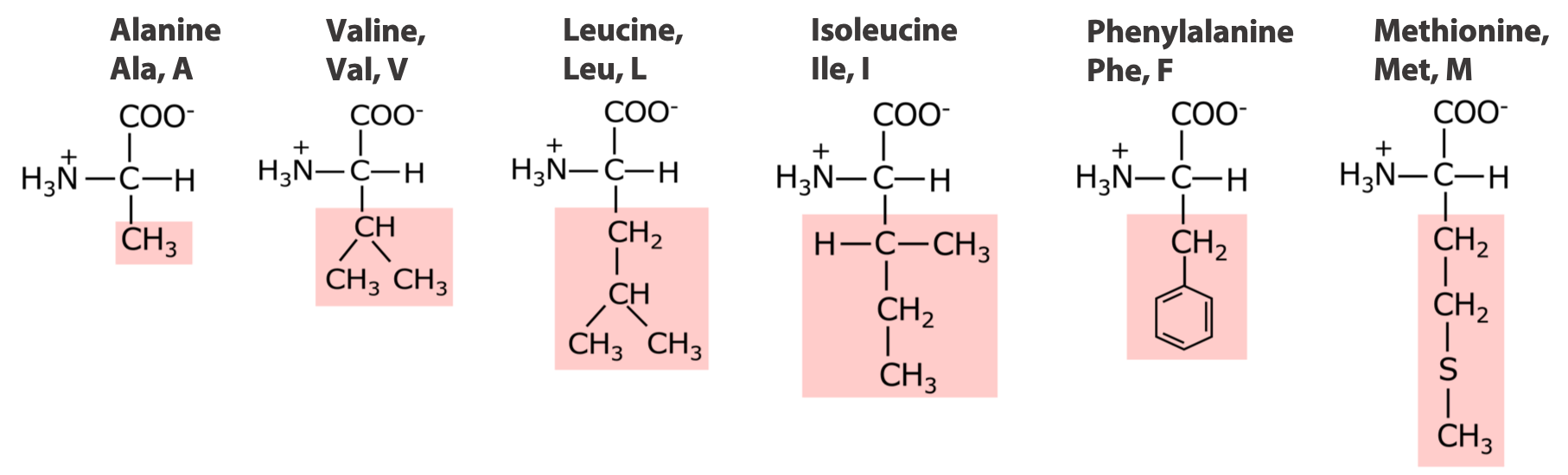
5 amino acids with medium to moderately non-polar side chains:
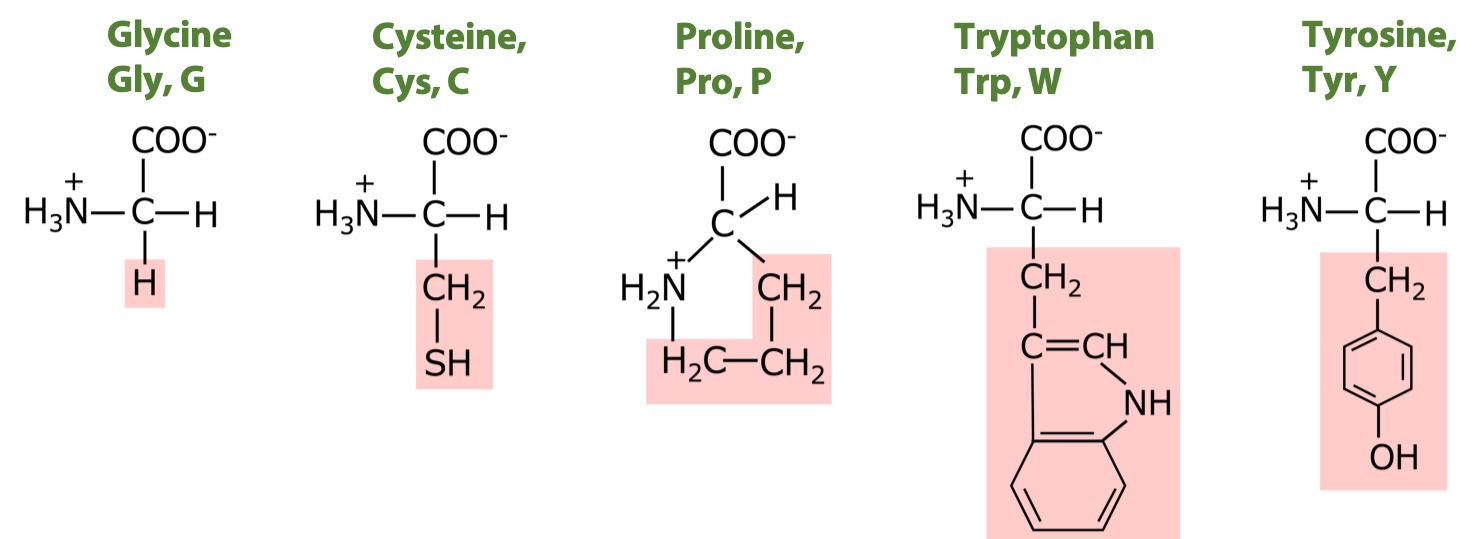
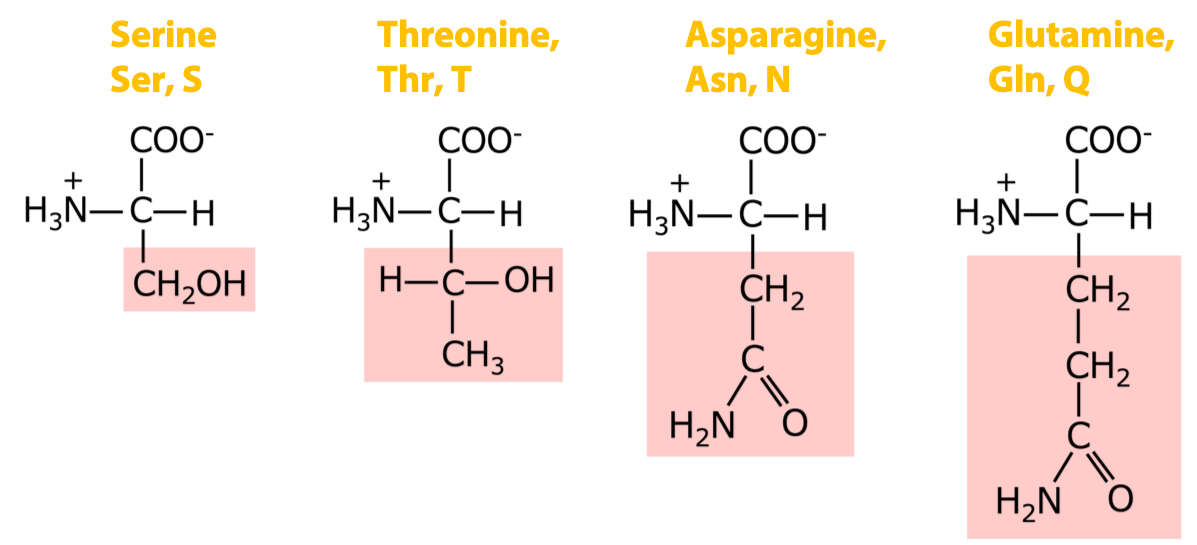
4 amino acids with polar uncharged side chains:
3 amino acids with positively charged side chains:
2 amino acids with negatively charged side chains:

Self-assessment Questions
