LAB 10: Food Lab
Learning Objectives
- Determine the number of aerobic bacteria, select pathogs, yeast, and mould in a food sample.
Introduction
In Canada, testing food for the presence of bacteria is regulated by the federal government. Health Canada sets out protocols for testing all bacteria (either aerobic or anaerobic) or specific pathogens.
In this lab, we will be following the protocol:
- MFHPB-18 Determination of the aerobic colony count in foods
Aerobic Colony Count
The food sample is homogenized then diluted water. Bacterial cells will recover better and grow on plates more often if they were diluted in peptone rather than water, but we are using water due to contamination issues with peptone. The bacteria on the food sample may be stressed and not in their optimal conditions, thus, if we are trying to count them in the lab, we will treat them as gently as possible to prevent killing these already stressed bacteria.
This protocol does not determine if the bacteria are pathogenic or merely normal flora on the food. The presence and number of spoilage bacteria depends on factors related to the food itself, called intrinsic factors, and factors related to the handling and storage of the food, called extrinsic factors.
- Refer to chapter 16 of the text for more information on food spoilage.
The peptone is then plated onto a non-selective rich medium for bacteria. The plates are incubated for 24 hours at 37 °C then counted.
The goal of MFHPB-18 is to ensure compliance with sections 4 and 7 of the Food and Drugs Act:
4. (1) No person shall sell an article of food that
(a) has in or on it any poisonous or harmful substance;
(b) is unfit for human consumption;
(c) consists in whole or in part of any filthy, putrid, disgusting, rotten, decomposed or diseased animal or vegetable substance;
(d) is adulterated; or
(e) was manufactured, prepared, preserved, packaged or stored under unsanitary conditions.
7. No person shall manufacture, prepare, preserve, package or store for sale any food under unsanitary conditions.
Xylose Lysine Desoxycholate Agar (XLD)
The previous method will determine total aerobic bacteria. Food manufactures are also required to test for specific pathogens. Selective and differential media is a common culture-based method to detect pathogens.
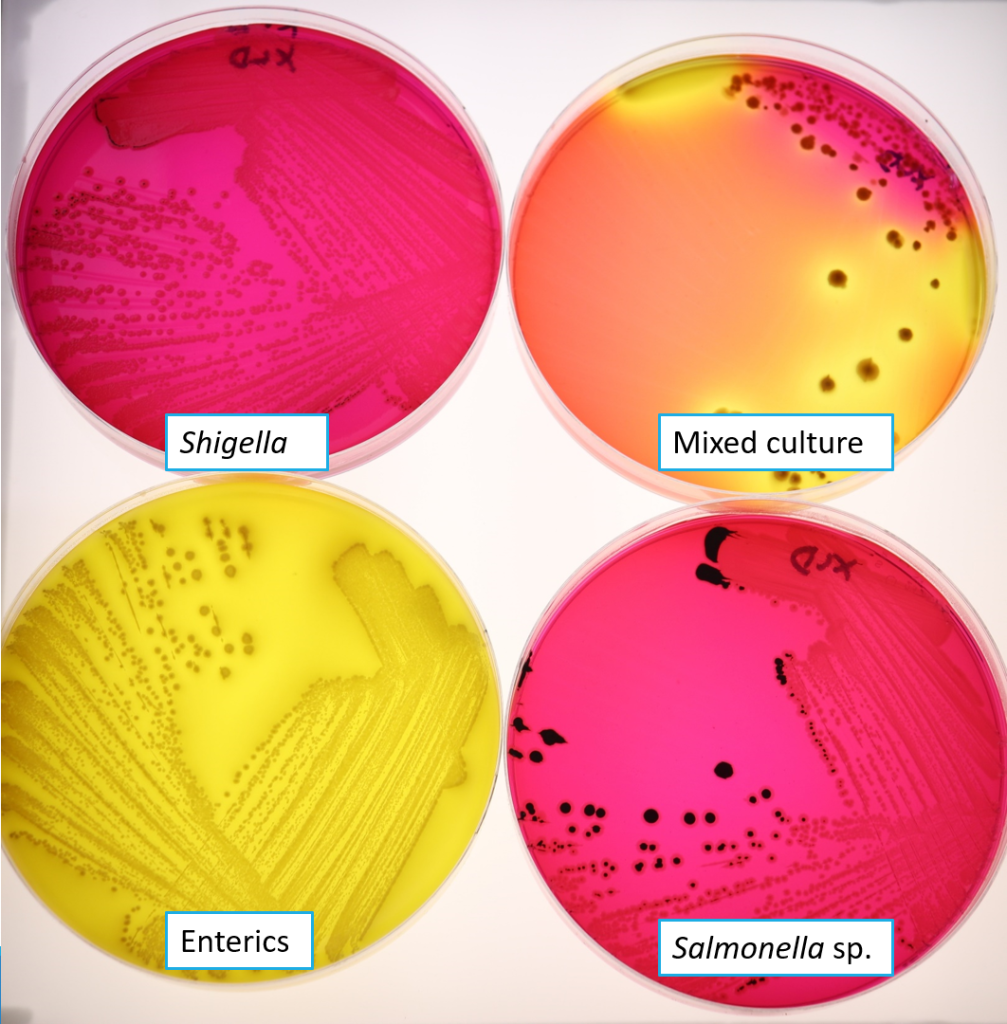
XLD is used for the detection of Salmonella and Shigella. The selective agent in XLD, desoxycholate, inhibits the growth of Gram positive bacteria. Xylose is the carbohydrate source that will turn the colonies red if it is fermented. Xylose fermentation is universal among enteric bacteria, except Shigella. The pH indicator is phenol red, which turns colonies yellow if the pH decreases due to xylose fermentation. A second differential agent, lysine, is used to differentiate Salmonella. When lysine is decarboxylated by Salmonella, the pH of the medium increases, causing the colonies to appear red (after the pH was acidic due to xylose fermentation). Sulphur source thiosulphate allows hydrogen sulphide formation under alkaline conditions, resulting in colonies with black centres. Typical colonies you may see on XLD are shown in Figure 10.1
Enterics such as Citrobacter, Proteus, E. coli: yellow colonies
- xylose fermented, lys not decarboxylated, hydrogen sulphide not produced
Salmonella: red colonies with black centres
- xylose fermentated, lys decarboxylated, hydrogen sulphide produced
Shigella: red colonies - xylose not fermented, lys not decarboxylated, hydrogen sulphide not produced
Yeast and Mould are often detected in food in addition to bacteria. The growth conditions for yeast and mould are often lower temperature than bacterial pathogens and longer incubations. We will use petri film and agar plates for the yeast and mould enumeration. Petrifilms are easy to use and less waste than a petri plate. These are often used in industry. Yeast and mould agar that we are using is potato dextrose agar (PDA). Chloramphenicol is added to inhibit bacterial growth, otherwise bacteria would overgrow the plate. The medium is named PDA-c to indicate the presence of chloramphenicol.
Listeria is found in the soil and untreated water. It contaminates meat, dairy products, fruits and vegetables. It also can contaminate processed foods if the processing facility is contaminated. Since listeriosis poses a significant risk to the elderly, pregnant people and those with weakened immune systems, Listeria are monitored in food processing facilities. We will be following a modified MFHPB-30 from Health Canada. This protocol involves enriching (increasing bacterial numbers) the sample, then plating on selective media. Listeria enrichment broth (LEB for short) allows Listeria spp. To multiply while inhibiting the growth of other common organisms through the action of antibiotics in the medium (cycloheximide inhibits fungi, nalidixic acid and acriflavine inhibit bacteria). The broth is then streaked onto Oxford Agar after enrichment. This agar is inhibitory to gram negatives and most gram positives using the selective agents lithium chloride, colisitin, cycloheximide, acriflavine and Fosfomycin. Listeria produces a black precipitate in the agar surrounding the coloines. Soil frequently contains Listeria, however it is often non-pathogenic L. innocua.
Staphylococcus aureus is a common source of food poisoning. There are many strains of this species, but the strains responsible for food poisoning produce enterotoxin. These strains are typically also coagulase positive. S. aureus the produce the coagulase enzyme are able to form a clot in plasma. Detecting the production of coagulase is a way to differentiate strains causing food poisoning from other strains.
We will be using Mannitol Salt Agar (MSA) as before. Coagulase-positive S. aureus produce yellow colonies with bright yellow zones. Coagulase-negative strains produce small red colonies with no change in colour to the agar surrounding them (recall the appearance of S. epidermidis on MSA). We are following Health Canada’s protocol MFHPB-21.
Bacillus cereus is a common contaminant in foods. When it grows to high densities, enough enterotoxin can be produced to cause food poisoning. Bacillus species make the antibiotic polymyxin and are resistant to it. We can select for Bacillus by growing in media with polymyxin. We will be using FDA protocol from the Bacteriological Analytical Manual Chapter 14, which is followed by Public Health Ontario. This method involves MPN tubes with selective media containing polymyxin B at 0.15% in trypticase soy broth. Turbidity is a positive result and the number of positive tubes is used to approximate the inoculum in the food source. You will do a confirmatory test by making a smear from the broth and Gram staining the culture. Bacillus cereus are Gram positive rods in short to long chains. Spores, if present, are ellipsoid-shaped and central to subterminal (around the middle of the cell).
Food Exercise
Materials
- 2 nutrient agar (NA or TSA) plates
- 2 petri film plate for yeast and mould
- 2 MSA plates
- 3 double strength TSB+polymyxin
- 6 single strength TSB+polymyxin
- 1 Listeria Enrichment Broth tube
- 1 10 ml pipette (2 if you have a liquid food sample)
- 6 microfuge tubes
- P100 pipette and yellow tips
- P1000 pipette and blue tips
- Sterile water
- Empty petri plate (if you have a solid sample)
- Scalpel (if you have a solid sample)
- Vortex
Method
Aerobic Colony Count
- Solid food: Remove several representative pieces off solid from different locations to get 10 g. Cut into small, uniform pieces in a sterile petri plate using a sterile scalpel.
Liquid food: Shake food to mix. Remove 10 ml using a pipette. - Add the sample to 90 ml 0.1% peptone. Shake vigorously for 30 seconds to mix.
- With a pipette, transfer 10 ml from the first bottle to the second bottle. Make sure both bottles were previously labeled with the dilution. Shake the second bottle as above.
- Your professor will instruct which dilutions to plate. For samples with more bacteria, you will need to do extra dilutions.
- Refer to the growth lab for the procedure on serial dilutions. An outline is below.
- DROP PLATING: We are going to perform the remaining 6 dilutions in microfuge tubes and put all 8 dilutions onto the same Petri plate. These are 10-fold serial dilutions like you did in the growth lab. Refer to the drop plating protocol in the growth lab.
Note: If there is no growth on the lowest dilution, represent the value as “<“.
Remember, there was a plating factor because 0.02 ml was plated for each dilution (instead of 1 ml being plated). Take this into account when calculating the dilution factor. Results are reported in cfu/ml.
XLD and MSA for pathogen detection
- From previously prepared dilutions (technique MFHPB-18), plate 100 µl onto each XLD and MSA. Do not spread right to the edge of the plate as confluent growth can occur along the edge of the plate, which is difficult to count.
- Bottle 1 (10-1 dilution) à two plates: one XLD, one MSA
- Bottle 2 (10-2 dilution) à one plate: one XLD, one
- Incubate at 37 ̊C for 18-24 hours for XLD and MSA. Colony counts can be verified at 48 hours if required.
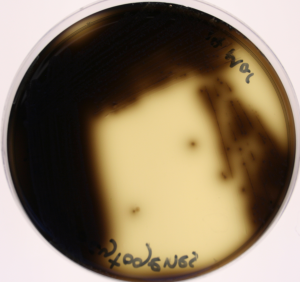
- On XLD: Count any Salmonella, Shigella and other enterics. If there are more than 150 colonies on the plate, report it as TNTC.
- On Baird-Parker, count any colonies that are 2-3 mm on an uncrowded plate, grey to jet black with an off-white margin.
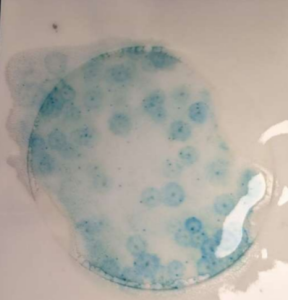
Petri Film for yeast and mould
- Lift the top film and place 1 ml of 10-2 dilution onto petrifilm rapid yeast and mould count plate. Repeat using 1 ml of 10-3 dilution.
- Roll the top film down.
- Place a flat object on the film to spread the sample volume across the film (an Erlenmeyer flask works well for this).
- Incubate at 25 C for 48 hours.
- Count the number of yeast and mould colonies that form. Yeast are small with a defined edge. Mould are large, flat with a diffuse edge.
MPN for Bacillus cereus enumeration
- Perform MPN test as was done in the water lab.
- Incubate tubes 48 hours at 30 C.
- Positive tubes are turbid. Consult the table in BAM Appendix 2: Most Probable Number from Serial Dilutions for determining the most probable number.
- Make a smear from one of the positive tubes.
- Perform a Gram stain on the smear. Take pictures to identify if the cell morphology matches B. cereus.
Listeria monocytogenes detection
- Cut sample into small pieces with sterile scalpel.
- Transfer 2 g of sample into 10 ml Listeria Enrichment Broth.
- Incubate at 37 C for 24 hours.
- Transfer 10 ul of broth to Oxford Agar plate.
- Streak plate as normal.
- Incubate at 37 C for 24 hours.
- Listeria colonies are 1 mm with a black halo in the agar. Colonies can also appear green-black or brown-black.
Staphylococcus aureus enumeration in food