9.3 Translation
Translation refers to the process by which a ribosome reads the genetic message in mRNA to synthesize a protein. This involves decoding the sequence of nucleotides in mRNA to assemble amino acids in the correct order, forming a protein.
Relating this to translating languages, think of mRNA as a message written in one language (nucleotides) that needs to be translated into another language (amino acids). Just as a translator may use a French-English dictionary to convert text from one language to another, the ribosome uses the genetic code to translate the sequence of nucleotides in mRNA into a sequence of amino acids to build a protein.
Genetic Code
The genetic code consists of the sequence of nitrogen bases in a chain of DNA or RNA. The bases are adenine (A), cytosine (C), guanine (G), and thymine (T) (or uracil, U, in RNA). The four bases make up the “letters” of the genetic code. The letters are combined in groups of 3 to form code “words,” called codons. Each codon stands for (encodes) one amino acid unless it codes for a start or stop signal. There are 20 common amino acids in proteins. With four bases forming three-base codons, there are 64 possible codons. This is more than enough to code for the 20 amino acids.
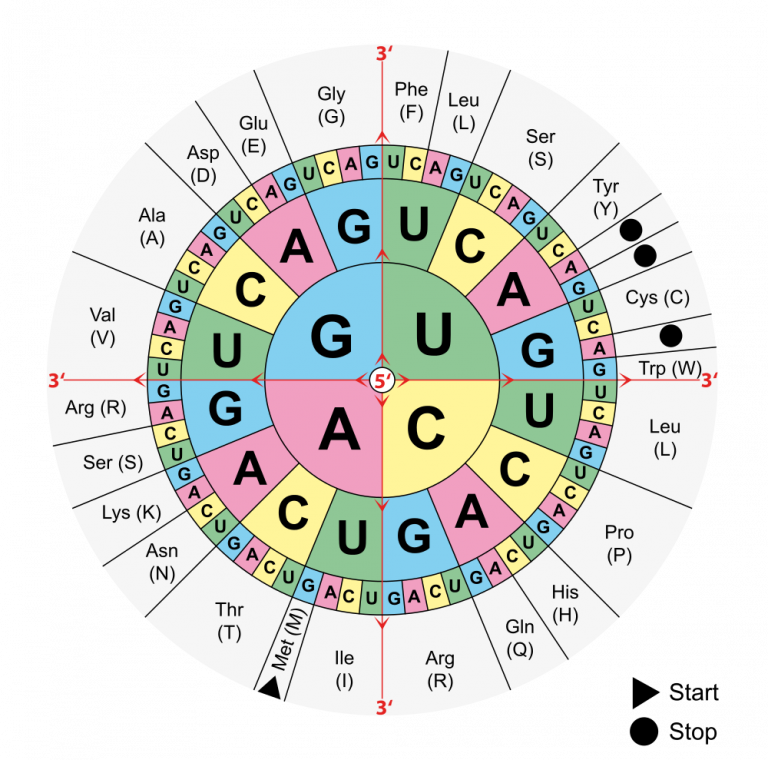
Figure 9.3.1 Description
Circular codon chart showing the genetic code. It is read from the center outward, starting with the first nucleotide of an mRNA codon at the center, followed by the second and third nucleotides. The chart maps mRNA codons to their corresponding amino acids, using one-letter and three-letter abbreviations. Start codon (AUG) is indicated for methionine (Met), and three-stop codons are marked with black dots.
To find the amino acid for a particular codon, find the first base in the codon in the circle’s centre in Figure 9.3.1, then the second base in the middle row out from the center, and finally, the third base in the outer ring. For example, CUG codes for leucine, AAG codes for lysine, and GGG codes for glycine.
If you find the codon AUG in Figure 9.3.1, you will see that it codes for the amino acid methionine. This codon is also the start codon that establishes the reading frame of the code. The start codon is a necessary tool in translation since a single chromosome contains many genes. To transcribe and translate a gene for a specific protein, we need to know where in the DNA code to start “reading” the instructions. AUG signals the start of a reading frame. After the AUG start codon, the next three bases are read as the second codon. The next three bases after that are read as the third codon, and so on. The sequence of bases is read, codon by codon until a stop codon is reached. UAG, UGA, and UAA are all stop codons. They do not code for any amino acids.
Exercise 9.3.1
Text Description
- 4
- 20
- 64
- It depends on the species.
5. How many common amino acids are there in proteins?
- 4
- 20
- 64
- It depends on the species.
6. Drop Zones
- Reading Frame
- Met
- Arg
- Pro
- Start Codon
Available Answers
- Start Codon
- Pro
- Arg
- Met
- Reading Frame
Translation
Translation is the second part of the central dogma of molecular biology: RNA → Protein.
It is the process in which the genetic code in mRNA is read to make a protein.
After mRNA leaves the nucleus, it moves to a ribosome, which consists of rRNA and proteins. The ribosome reads the sequence of codons in mRNA, and tRNA molecules bring amino acids to the ribosome in the correct sequence.
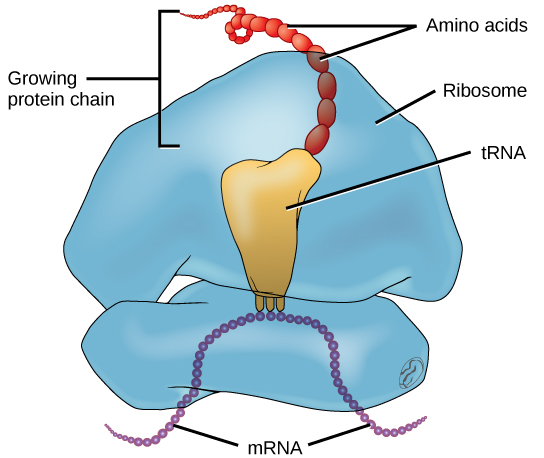
Figure 9.3.2 Description
Diagram of translation showing a ribosome bound to mRNA. A tRNA molecule delivers amino acids to the ribosome, where they are assembled into a growing protein chain. The mRNA strand is threaded through the ribosome, guiding the sequence of amino acids.
Translation occurs in three stages:
1. Initiation
After transcription in the nucleus, the mRNA exits through a nuclear pore and enters the cytoplasm. At the region on the mRNA containing the cap and the start codon, the small and large subunits of the ribosome bind to the mRNA. These are then joined by a tRNA, which contains the anticodons that match the start codon (start signal) on the mRNA. This group of molecules (mRNA, ribosome, tRNA) is called an initiation complex.
2. Elongation
tRNA keeps bringing amino acids to the growing polypeptide according to complementary base pairing between the codons on the mRNA and the anticodons on the tRNA. As a tRNA moves into the ribosome, its amino acid is transferred to the growing polypeptide. Once this transfer is complete, the tRNA leaves the ribosome, the ribosome moves one codon length down the mRNA, and a new tRNA enters with its corresponding amino acid. This process repeats, and the polypeptide grows.
3. Termination
At the end of the mRNA coding is a stop codon (stop signal), which will end the elongation stage. The stop codon doesn’t call for a tRNA but instead for a type of protein called a release factor, which will cause the entire complex (mRNA, ribosome, tRNA, and polypeptide) to break apart, releasing all components.
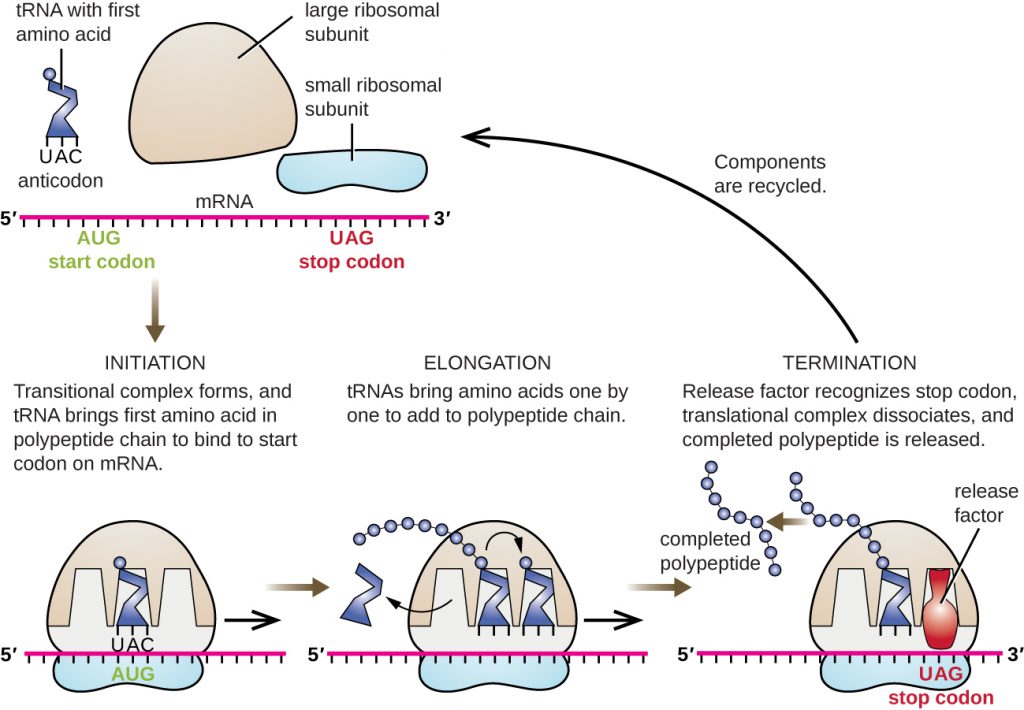
Figure 9.3.3 Description
Diagram of the three translation stages in protein synthesis: initiation, elongation, and termination. At initiation, a tRNA with the anticodon UAC binds to the start codon AUG on the mRNA, forming a complex with the small and large ribosomal subunits. During elongation, additional tRNAs bring amino acids to the ribosome, which are linked to form a polypeptide chain. In termination, the ribosome reaches a stop codon (UAG), and a release factor binds, releasing the completed polypeptide and allowing the ribosomal components to be recycled.
Exercise 9.3.2
Text Description
DNA is copied onto a strand of mRNA during a process called ____.
The code on mRNA is “read” and used to create a protein during a process called ____.
This process is further broken down into three stages;
In ____ the ribosome, mRNA and first tRNA all associate.
In ____ amino acids are brought to the ribosome by tRNA and are added to the growing protein.
In ____ a stop codon is reached, and the complex dissociates and released the newly formed protein.
____ contains anticodons.
____ makes up the ribosome along with proteins.
– non-template strand will read ____
– mRNA codons will read ____
– the tRNA anticodons matching the mRNA will read: ____
- DNA
- mRNA
- tRNA
- Ribosomes
Correct answer(s):DNA is copied onto a strand of mRNA during a process called transcription.
The code on mRNA is “read” and used to create a protein during a process called translation.
This process is further broken down into three stages;
In initiation the ribosome, mRNA and first tRNA all associate.
In elongation amino acids are brought to the ribosome by tRNA and are added to the growing protein.
In termination a stop codon is reached, and the complex dissociates and released the newly formed protein.
2.
tRNA contains anticodons.
rRNA makes up the ribosome along with proteins.
Correct answer(s):If a sequence of DNA on the template strand reads CAG TAC, then the:
– non-template strand will read GTC ATG
– mRNA codons will read GUC AUG
– the tRNA anticodons matching the mRNA will read: CAG UAC
Mutations
Mutations are random changes in the sequence of bases in DNA or RNA.
Mutations have many possible causes. Some mutations seem to happen spontaneously, without any outside influence. They occur when errors are made during DNA replication or during the transcription phase of protein synthesis. Environmental factors cause other mutations. Anything in the environment that can cause a mutation is known as a mutagen.
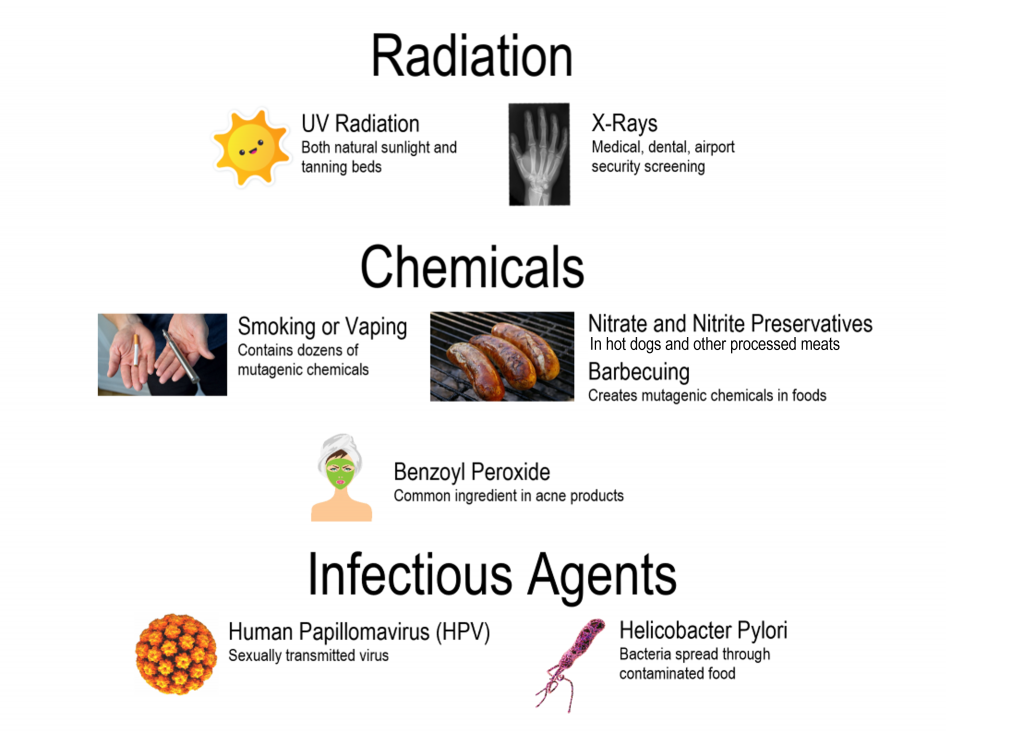
Figure 9.3.4 Description
Diagram categorizing common mutagens into three groups: Radiation, Chemicals, and Infectious Agents. Under Radiation: UV Radiation (from sunlight and tanning beds) and X-rays (used in medical, dental, and airport screenings). Under Chemicals: Smoking or Vaping (contains mutagenic chemicals), Nitrate and Nitrite Preservatives and Barbecuing (create mutagenic compounds in food), and Benzoyl Peroxide (an acne treatment ingredient). Under Infectious Agents: Human Papillomavirus (HPV), a sexually transmitted virus, and Helicobacter Pylori, a bacterium spread through contaminated food.
Types of Mutations
There are many different types of mutations:
Chromosomal Alterations
Chromosomal Alterations are mutations that change chromosome structure. They occur when a section of a chromosome breaks off and rejoins incorrectly, or otherwise does not rejoin at all. Possible ways in which these mutations can occur are illustrated in the figure below. Chromosomal alterations are very serious. They often result in the death of the organism in which they occur.
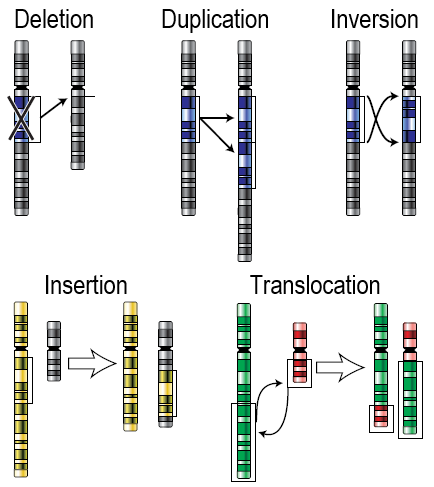
Figure 9.3.5 Description
Diagram illustrating five types of chromosomal mutations: Deletion (a segment is removed), Duplication (a segment is copied), Inversion (a segment is reversed), Insertion (a segment from one chromosome is inserted into another), and Translocation (segments are exchanged between non-homologous chromosomes). Each mutation type is shown with arrows indicating the change in chromosome structure.
Point Mutation
A point mutation is a change in a single nucleotide in DNA. This type of mutation is usually less serious than a chromosomal alteration. An example of a point mutation is one that changes the codon UUU to the codon UCU. Point mutations can be silent, missense, or nonsense mutations, as described in the table below. The effects of point mutations depend on how they change the genetic code.
|
Type |
Description |
Example |
Effect |
|---|---|---|---|
| Silent | Mutated codon codes for the same amino acid | CAA (glutamine) → CAG (glutamine) | None |
| Missense | Mutated codon codes for a different amino acid | CAA (glutamine) → CCA (proline) | Variable |
| Nonsense | Mutated codon is a premature stop codon | CAA (glutamine) → UAA (stop) | Usually serious |
Frameshift Mutation
A frameshift mutation is a deletion or insertion of one or more nucleotides, changing the reading frame of the base sequence.
The reading frame refers to how the sequence of RNA bases is divided into codons (groups of three nucleotides that each code for a specific amino acid). The ribosome reads the mRNA three bases at a time, starting from the start codon (AUG). If a nucleotide is added or removed, the grouping of the bases shifts, and every codon after the mutation is read incorrectly.
Think of the reading frame like spacing in a sentence: “THE CAT ATE THE RAT”. If you insert one letter, then it changes to “TTH ECA TAT ETH ERA T”. Even though most of the letters are the same, the meaning is completely lost.
Consider the following sequence of bases in RNA:
AUG-AAU-ACG-GCU = start-asparagine-threonine-alanine
Now, assume that an insertion occurs in this sequence. Let’s say an A nucleotide is added after the start codon AUG. The sequence of bases becomes:
AUG-AAA-UAC-GGC-U = start-lysine-tyrosine-glycine
Even though the rest of the sequence is unchanged, this insertion shifts the reading frame and alters all the codons that follow. As this example shows, a frameshift mutation can dramatically change how the codons in mRNA are read, often resulting in a completely different and nonfunctional protein.
Effects of Mutations
Everyone has mutations. In fact, most people have dozens (or even hundreds!) of mutations in their DNA. From an evolutionary perspective, mutations are essential. They are needed for evolution to occur because they are the ultimate source of all new genetic variation in any species.
Most mutations have neither negative nor positive effects on the organism in which they occur. These mutations are called neutral mutations. Examples include silent point mutations, which are neutral because they do not change the amino acids in the proteins they encode. Many other mutations do not affect the organism because they are repaired before protein synthesis occurs. Cells have multiple repair mechanisms to fix mutations in DNA.
Some mutations — known as beneficial mutations — have a positive effect on the organism in which they occur. They generally code for new versions of proteins that help organisms adapt to their environment. If they increase an organism’s chances of surviving or reproducing, the mutations will likely become more common over time. For example, mutations allow some bacteria to survive in the presence of antibiotic drugs, leading to the evolution of antibiotic-resistant strains of bacteria. Mutations are needed for evolution because they create genetic differences, which help species adapt and change over time.
Imagine making a random change in a complicated machine, such as a car engine. There is a chance that the random change would result in a car that does not run well — or perhaps does not run at all. By the same token, a random change in a gene’s DNA may result in the production of a protein that does not function normally… or may not function at all. Such mutations are likely to be harmful. Harmful mutations may cause genetic disorders or cancer.
Mutations and Cancer
Some types of cancer occur because of gene mutations that control the cell cycle. Cancer-causing mutations most often occur in two types of regulatory genes: proto-oncogenes and tumor-suppressor genes.
Proto-oncogenes are genes that normally help cells divide. When a proto-oncogene mutates to become an oncogene, it is continuously expressed, even when it is not supposed to be. This is like a car’s accelerator pedal being stuck at full throttle. The car keeps racing at top speed. A cell, in this case, keeps dividing out of control, which can lead to cancer.
Tumour suppressor genes are genes that normally slow down or stop cell division. When a mutation occurs in a tumour suppressor gene, it can no longer control cell division. This is like a car without brakes. The car can’t be slowed or stopped. A cell, in this case, keeps dividing out of control, which can lead to cancer.
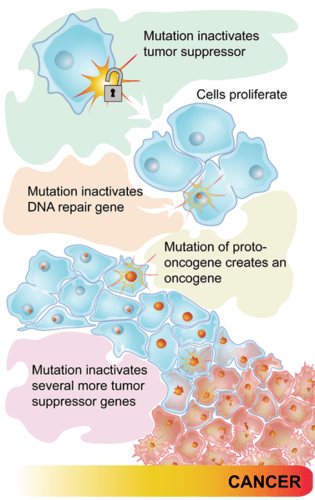
A series of mutations are often required in the development of cancer. Mutations must convert proto-oncogenes into oncogenes to trigger uncontrolled cell growth. Then tumour suppressor genes must be inactivated to allow continued growth, eventually leading to the formation of cancerous cells.
Figure 9.3.6 Description
Diagram showing the stepwise development of cancer through genetic mutations. It begins with a mutation that inactivates a tumour suppressor gene, allowing cells to proliferate. A second mutation inactivates a DNA repair gene. A third mutation converts a proto-oncogene into an oncogene, further promoting cell growth. Additional mutations inactivate more tumour suppressor genes, eventually leading to the formation of cancerous cells.
Exercise 9.3.3
Text Description
- Frameshift
- Chromosomal
- Inversion
- Point
- False
- True
- Point
“9.4 Translation” from Biology and the Citizen by Colleen Jones is licensed under a Creative Commons Attribution 4.0 International License, except where otherwise noted.
“5.6 Genetic Code” from Human Biology by Christine Miller is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, except where otherwise noted.
“5.8 Mutations” from Human Biology by Christine Miller is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, except where otherwise noted.

